+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1hhu | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Balhimycin in complex with D-Ala-D-Ala | |||||||||||||||
 Components Components | BALHIMYCIN | |||||||||||||||
 Keywords Keywords | ANTIBIOTIC / GLYCOPEPTIDE / CELL WALL PEPTIDES | |||||||||||||||
| Function / homology | Balhimycin / beta-D-glucopyranose / CITRIC ACID / D-ALANINE / Chem-DVC / :  Function and homology information Function and homology information | |||||||||||||||
| Biological species |  AMYCOLATOPSIS SP. (bacteria) AMYCOLATOPSIS SP. (bacteria) | |||||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / DIRECT METHODS / Resolution: 0.89 Å SYNCHROTRON / DIRECT METHODS / Resolution: 0.89 Å | |||||||||||||||
 Authors Authors | Lehmann, C. / Bunkoczi, G. / Sheldrick, G.M. / Vertessy, L. | |||||||||||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2002 Journal: J.Mol.Biol. / Year: 2002Title: Structures of Glycopeptide Antibiotics with Peptides that Model Bacterial Cell-Wall Precursors Authors: Lehmann, C. / Bunkoczi, G. / Vertesy, L. / Sheldrick, G.M. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1hhu.cif.gz 1hhu.cif.gz | 53.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1hhu.ent.gz pdb1hhu.ent.gz | 44.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1hhu.json.gz 1hhu.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hh/1hhu https://data.pdbj.org/pub/pdb/validation_reports/hh/1hhu ftp://data.pdbj.org/pub/pdb/validation_reports/hh/1hhu ftp://data.pdbj.org/pub/pdb/validation_reports/hh/1hhu | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 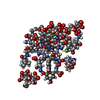
| ||||||||||||||||
| 2 | 
| ||||||||||||||||
| Unit cell |
| ||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
|
- Components
Components
-Protein/peptide , 1 types, 4 molecules ABCD
-Sugars , 2 types, 8 molecules 


| #2: Sugar | ChemComp-BGC /   Type: D-saccharide, beta linking, Glycopeptide / Class: Antibiotic, Antimicrobial / Mass: 180.156 Da / Num. of mol.: 4 Type: D-saccharide, beta linking, Glycopeptide / Class: Antibiotic, Antimicrobial / Mass: 180.156 Da / Num. of mol.: 4Source method: isolated from a genetically manipulated source Formula: C6H12O6 Details: BALHIMYCIN IS A TRICYCLIC HEPTAPEPTIDE GLYCOSYLATED BY D-GLUCOSE (RESIDUE 8) ON RESIDUE 4 AND BY 4-OXO-VANCOSAMINE (RESIDUE 9) ON RESIDUE 6. References: Balhimycin #3: Sugar | ChemComp-DVC / (   Type: L-saccharide, alpha linking, Glycopeptide / Class: Antibiotic, Antimicrobial / Mass: 177.198 Da / Num. of mol.: 4 Type: L-saccharide, alpha linking, Glycopeptide / Class: Antibiotic, Antimicrobial / Mass: 177.198 Da / Num. of mol.: 4Source method: isolated from a genetically manipulated source Formula: C7H15NO4 Details: BALHIMYCIN IS A TRICYCLIC HEPTAPEPTIDE GLYCOSYLATED BY D-GLUCOSE (RESIDUE 8) ON RESIDUE 4 AND BY 4-OXO-VANCOSAMINE (RESIDUE 9) ON RESIDUE 6. References: Balhimycin |
|---|
-Non-polymers , 4 types, 140 molecules 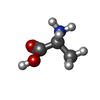






| #4: Chemical | ChemComp-DAL / #5: Chemical | ChemComp-CIT / #6: Chemical | #7: Water | ChemComp-HOH / | |
|---|
-Details
| Compound details | BALHIMYCIN IS A TRICYCLIC GLYCOPEPTIDE. THE SCAFFOLD IS A HEPTAPEPTIDE WITH THE CONFIGURATION D-D-L- ...BALHIMYCIN |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.14 Å3/Da / Density % sol: 22.4 % |
|---|---|
| Crystal grow | pH: 7 / Details: 1M CIT, PH =7, 25% MPD |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  EMBL/DESY, HAMBURG EMBL/DESY, HAMBURG  / Beamline: X11 / Wavelength: 0.9076 / Beamline: X11 / Wavelength: 0.9076 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Sep 15, 1998 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9076 Å / Relative weight: 1 |
| Reflection | Resolution: 0.89→27.99 Å / Num. obs: 42306 / % possible obs: 99.4 % / Redundancy: 3.66 % / Rmerge(I) obs: 0.0169 / Net I/σ(I): 38.76 |
| Reflection shell | Resolution: 0.89→1 Å / Redundancy: 3.35 % / Rmerge(I) obs: 0.0339 / Mean I/σ(I) obs: 24.67 / % possible all: 98.1 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: DIRECT METHODS / Resolution: 0.89→27.99 Å / Num. parameters: 6200 / Num. restraintsaints: 9467 / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: ENGH AND HUBER / Details: FRIDEL OPPOSITES DURING REFINEMENT NOT MERGED
| |||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: METHOD USED: MOEWS & KRETSINGER, J.MOL.BIOL.91(1973)201-228 | |||||||||||||||||||||||||||||||||
| Refine analyze | Num. disordered residues: 28 / Occupancy sum hydrogen: 354.5 / Occupancy sum non hydrogen: 630.5 | |||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 0.89→27.99 Å
| |||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj







