[English] 日本語
 Yorodumi
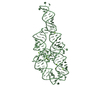
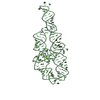
Yorodumi- PDB-1gid: CRYSTAL STRUCTURE OF A GROUP I RIBOZYME DOMAIN: PRINCIPLES OF RNA... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1gid | ||||||
|---|---|---|---|---|---|---|---|
| Title | CRYSTAL STRUCTURE OF A GROUP I RIBOZYME DOMAIN: PRINCIPLES OF RNA PACKING | ||||||
 Components Components | P4-P6 RNA RIBOZYME DOMAIN | ||||||
 Keywords Keywords | RIBOZYME / RNA / P4-P6 RIBOZYME DOMAIN OF THE TETRAHYMENA GROUP I INTRON | ||||||
| Function / homology | COBALT HEXAMMINE(III) / RNA / RNA (> 10) / RNA (> 100) Function and homology information Function and homology information | ||||||
| Biological species | synthetic construct (others) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / MAD-SIR / Resolution: 2.5 Å SYNCHROTRON / MAD-SIR / Resolution: 2.5 Å | ||||||
 Authors Authors | Cate, J.H. / Gooding, A.R. / Podell, E. / Zhou, K. / Golden, B.L. / Kundrot, C.E. / Cech, T.R. / Doudna, J.A. | ||||||
 Citation Citation |  Journal: Science / Year: 1996 Journal: Science / Year: 1996Title: Crystal structure of a group I ribozyme domain: principles of RNA packing. Authors: Cate, J.H. / Gooding, A.R. / Podell, E. / Zhou, K. / Golden, B.L. / Kundrot, C.E. / Cech, T.R. / Doudna, J.A. #1:  Journal: Science / Year: 1996 Journal: Science / Year: 1996Title: RNA Tertiary Structure Mediation by Adenosine Authors: Cate, J.H. / Gooding, A.R. / Podell, E. / Zhou, K. / Golden, B.L. / Szewczak, A.A. / Kundrot, C.E. / Cech, T.R. / Doudna, J.A. #2:  Journal: Structure / Year: 1996 Journal: Structure / Year: 1996Title: Metal-Binding Sites in the Major Groove of a Large Ribozyme Domain Authors: Cate, J.H. / Doudna, J.A. | ||||||
| History |
| ||||||
| Remark 999 | SEQUENCE REFERENCE: MURPHY, F.L., CECH, T.R. BIOCHEMISTRY 32, 5291 (1993). |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1gid.cif.gz 1gid.cif.gz | 182.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1gid.ent.gz pdb1gid.ent.gz | 138.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1gid.json.gz 1gid.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1gid_validation.pdf.gz 1gid_validation.pdf.gz | 365.1 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1gid_full_validation.pdf.gz 1gid_full_validation.pdf.gz | 394.3 KB | Display | |
| Data in XML |  1gid_validation.xml.gz 1gid_validation.xml.gz | 9.7 KB | Display | |
| Data in CIF |  1gid_validation.cif.gz 1gid_validation.cif.gz | 15.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gi/1gid https://data.pdbj.org/pub/pdb/validation_reports/gi/1gid ftp://data.pdbj.org/pub/pdb/validation_reports/gi/1gid ftp://data.pdbj.org/pub/pdb/validation_reports/gi/1gid | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: RNA chain | Mass: 51051.324 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: T7 TRANSCRIPT / Source: (natural) synthetic construct (others) #2: Chemical | ChemComp-MG / #3: Chemical | ChemComp-NCO / #4: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.3 Å3/Da / Density % sol: 71 % | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Method: vapor diffusion / pH: 6 / Details: pH 6.00, VAPOR DIFFUSION | ||||||||||||||||||||||||||||||||||||||||
| Components of the solutions |
| ||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS | ||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 110 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  CHESS CHESS  / Beamline: A1 / Beamline: A1 |
| Detector | Detector: CCD / Date: Dec 30, 1995 |
| Radiation | Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Relative weight: 1 |
| Reflection | Resolution: 2.5→18 Å / Num. obs: 36401 / % possible obs: 73 % / Observed criterion σ(I): 2 / Redundancy: 4 % / Rsym value: 0.043 / Net I/σ(I): 23.7 |
| Reflection shell | Resolution: 2.77→2.86 Å / Redundancy: 2.7 % / Mean I/σ(I) obs: 4.7 / Rsym value: 0.264 / % possible all: 80.2 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: MAD-SIR / Resolution: 2.5→8 Å / Cross valid method: FREE R / σ(F): 2 Details: NUCLEIC ACID RNA-DNA PARAMETER FILE: G. PARKINSON,ET AL. (1996) ACTA CRYST. D52, 57-64
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 45 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine Biso |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.5→8 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | NCS model details: 14 GROUPS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.8→2.87 Å / Total num. of bins used: 20
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file | Serial no: 2 / Param file: DNA-RNA-MULTI-ENDO.PARAM / Topol file: DNA-RNA-MULTI-ENDO.TOP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.843 / Classification: refinement X-PLOR / Version: 3.843 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 2.5 Å / Lowest resolution: 8 Å / σ(F): 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 45 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor Rfree: 0.51 / % reflection Rfree: 5 % / Total num. of bins used: 20 |
 Movie
Movie Controller
Controller








 PDBj
PDBj


































