[English] 日本語
 Yorodumi
Yorodumi- PDB-1fiz: THREE DIMENSIONAL STRUCTURE OF BETA-ACROSIN FROM BOAR SPERMATOZOA -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1fiz | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | THREE DIMENSIONAL STRUCTURE OF BETA-ACROSIN FROM BOAR SPERMATOZOA | |||||||||
 Components Components | (BETA-ACROSIN ...) x 2 | |||||||||
 Keywords Keywords | HYDROLASE / anti-parallel beta-barrel | |||||||||
| Function / homology |  Function and homology information Function and homology informationacrosin / acrosomal matrix / acrosome reaction / fucose binding / amidase activity / D-mannose binding / single fertilization / activation of adenylate cyclase activity / serine-type peptidase activity / serine-type endopeptidase activity ...acrosin / acrosomal matrix / acrosome reaction / fucose binding / amidase activity / D-mannose binding / single fertilization / activation of adenylate cyclase activity / serine-type peptidase activity / serine-type endopeptidase activity / protein-containing complex / proteolysis Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 2.9 Å SYNCHROTRON / Resolution: 2.9 Å | |||||||||
 Authors Authors | Tranter, R. / Read, J.A. / Jones, R. / Brady, R.L. | |||||||||
 Citation Citation |  Journal: Structure Fold.Des. / Year: 2000 Journal: Structure Fold.Des. / Year: 2000Title: Effector sites in the three-dimensional structure of mammalian sperm beta-acrosin. Authors: Tranter, R. / Read, J.A. / Jones, R. / Brady, R.L. #1:  Journal: Thesis Journal: ThesisTitle: Three dimensional structure of beta-acrosin from ram and boar spermatozoa Authors: Tranter, R. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1fiz.cif.gz 1fiz.cif.gz | 72.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1fiz.ent.gz pdb1fiz.ent.gz | 52.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1fiz.json.gz 1fiz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fi/1fiz https://data.pdbj.org/pub/pdb/validation_reports/fi/1fiz ftp://data.pdbj.org/pub/pdb/validation_reports/fi/1fiz ftp://data.pdbj.org/pub/pdb/validation_reports/fi/1fiz | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 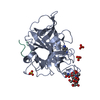
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Details | heterodimer of chain A and B linked together by two disulphide bonds |
- Components
Components
-BETA-ACROSIN ... , 2 types, 2 molecules AL
| #1: Protein | Mass: 29229.842 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #2: Protein/peptide | Mass: 2615.991 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-Sugars , 1 types, 1 molecules
| #3: Polysaccharide | beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1- ...beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source |
|---|
-Non-polymers , 3 types, 30 molecules 
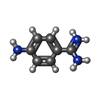



| #4: Chemical | ChemComp-SO4 / #5: Chemical | ChemComp-PBZ / | #6: Water | ChemComp-HOH / | |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.92 Å3/Da / Density % sol: 57.81 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop / pH: 6.5 Details: PEG 8000, ammonium sulphate, sodium cacodylate, p-aminobenzamidine, pH 6.5, VAPOR DIFFUSION, HANGING DROP, temperature 18K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS pH: 8.2 / Method: vapor diffusion | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SRS SRS  / Beamline: PX14.1 / Wavelength: 1.244 / Beamline: PX14.1 / Wavelength: 1.244 |
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD / Date: Feb 3, 2000 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.244 Å / Relative weight: 1 |
| Reflection | Resolution: 2.9→100 Å / Num. all: 8303 / Num. obs: 8303 / % possible obs: 97.4 % / Observed criterion σ(I): -3 / Redundancy: 5.9 % / Biso Wilson estimate: 65.8 Å2 / Rmerge(I) obs: 0.078 / Net I/σ(I): 18.6 |
| Reflection shell | Resolution: 2.9→3.1 Å / Redundancy: 5.9 % / Rmerge(I) obs: 0.307 / Num. unique all: 1200 / % possible all: 99.8 |
| Reflection | *PLUS Num. obs: 8711 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2.9→30 Å / SU B: 18.1713 / SU ML: 0.34735 / Cross valid method: THROUGHOUT / σ(F): 0 / σ(I): 2 / ESU R Free: 0.41073 Details: maximun likelihood refinement CNS was also used for refinement.
| ||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 47.027 Å2 | ||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.9→30 Å
| ||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: REFMAC / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 2.9 Å / Lowest resolution: 30 Å / σ(F): 0 / % reflection Rfree: 4.7 % | ||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller













 PDBj
PDBj

