[English] 日本語
 Yorodumi
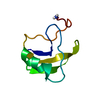
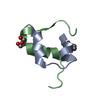
Yorodumi- PDB-1fh3: NMR STRUCTURES OF LQH III ALPHA-LIKE SCORPION TOXIN FROM LEIURUS ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1fh3 | ||||||
|---|---|---|---|---|---|---|---|
| Title | NMR STRUCTURES OF LQH III ALPHA-LIKE SCORPION TOXIN FROM LEIURUS QUINQUESTRIATUS CORRESPONDING TO THE MAJOR CONFORMER IN SOLUTION | ||||||
 Components Components | LQH III ALPHA-LIKE TOXIN | ||||||
 Keywords Keywords | TOXIN / ALPHA-LIKE TOXIN / SCORPION TOXIN / SODIUM CHANNEL INHIBITOR NON-PROLINE CIS PEPTIDE BOND / CIS-TRANS ISOMERISM | ||||||
| Function / homology |  Function and homology information Function and homology informationsodium channel inhibitor activity / defense response / toxin activity / extracellular region Similarity search - Function | ||||||
| Biological species |  Leiurus quinquestriatus hebraeus (scorpion) Leiurus quinquestriatus hebraeus (scorpion) | ||||||
| Method | SOLUTION NMR / distance geometry simulated annealing, molecular dynamics | ||||||
 Authors Authors | Krimm, I. / Trivelli, X. / Lancelin, J.M. | ||||||
 Citation Citation |  Journal: To be Published Journal: To be PublishedTitle: A Cis-trans Isomerism of a Non-prolyl Peptide Bond in Lqh III Alpha-like Scorpion Toxin Revealed by Solution NMR Authors: Krimm, I. / Trivelli, X. / Lancelin, J.M. #1:  Journal: J.Mol.Biol. / Year: 1999 Journal: J.Mol.Biol. / Year: 1999Title: NMR Structures and Activity of a Novel Alpha-like Toxin from the Scorpion Leiurus Quinquestriatus hebraeus Authors: Krimm, I. / Gilles, N. / Sautiere, P. / Stankiewicz, M. / Pelhate, M. / Gordon, D. / Lancelin, J.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1fh3.cif.gz 1fh3.cif.gz | 764.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1fh3.ent.gz pdb1fh3.ent.gz | 641 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1fh3.json.gz 1fh3.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fh/1fh3 https://data.pdbj.org/pub/pdb/validation_reports/fh/1fh3 ftp://data.pdbj.org/pub/pdb/validation_reports/fh/1fh3 ftp://data.pdbj.org/pub/pdb/validation_reports/fh/1fh3 | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
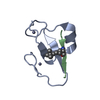
| #1: Protein | Mass: 7063.992 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: AMIDATED AT THE C-TERMINAL POSITION(CONH2). THIS STRUCTURE IS IN EQUILIBRIUM WITH THE PRO9-GLU10 TRANS ISOMER (1BMR). Source: (natural)  Leiurus quinquestriatus hebraeus (scorpion) Leiurus quinquestriatus hebraeus (scorpion)Secretion: VENOM / Species: Leiurus quinquestriatus / Strain: hebraeus / References: UniProt: P56678 |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 2mM LqhIII; 100 mM phosphate buffer pH 5.8; 90% H2O 10% D2O Solvent system: 90% H2O/10% D2O |
|---|---|
| Sample conditions | Ionic strength: NA / pH: 5.8 / Pressure: 1 atm / Temperature: 311 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Bruker AVANCE / Manufacturer: Bruker / Model: AVANCE / Field strength: 500 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: distance geometry simulated annealing, molecular dynamics Software ordinal: 1 | ||||||||||||||||||||
| NMR representative | Selection criteria: closest to the average | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with acceptable covalent geometry,structures with favorable non-bond energy,structures with the least restraint violations,structures with the lowest energy Conformers calculated total number: 80 / Conformers submitted total number: 42 |
 Movie
Movie Controller
Controller









 PDBj
PDBj

 X-PLOR
X-PLOR