[English] 日本語
 Yorodumi
Yorodumi- PDB-1feq: NMR SOLUTION STRUCTURE OF THE ANTICODON OF TRNA(LYS3) WITH T6A MO... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1feq | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | NMR SOLUTION STRUCTURE OF THE ANTICODON OF TRNA(LYS3) WITH T6A MODIFICATION AT POSITION 37 | ||||||||||||||||||
 Components Components | 5'-R(* Keywords KeywordsRNA / TRNA / Anticodon STEM LOOP / TRNA DOMAIN / RNA HAIRPIN / T6A | Function / homology | RNA / RNA (> 10) |  Function and homology information Function and homology informationMethod | SOLUTION NMR / global fold by distance geometry. Refinement by simulated annealing using the AMBER forcefield |  Authors AuthorsStuart, J.W. / Gdaniec, Z. / Guenther, R.H. / Marszalek, M. / Sochacka, E. / Malkiewicz, A. / Agris, P.F. |  Citation Citation Journal: Biochemistry / Year: 2000 Journal: Biochemistry / Year: 2000Title: Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A. Authors: Stuart, J.W. / Gdaniec, Z. / Guenther, R. / Marszalek, M. / Sochacka, E. / Malkiewicz, A. / Agris, P.F. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1feq.cif.gz 1feq.cif.gz | 124.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1feq.ent.gz pdb1feq.ent.gz | 101.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1feq.json.gz 1feq.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fe/1feq https://data.pdbj.org/pub/pdb/validation_reports/fe/1feq ftp://data.pdbj.org/pub/pdb/validation_reports/fe/1feq ftp://data.pdbj.org/pub/pdb/validation_reports/fe/1feq | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 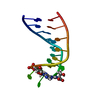
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 5510.319 Da / Num. of mol.: 1 Fragment: ANTICODON (RESIDUES 27-43) OF TRNA(LYS3) WITH T6A MODIFICATION AT POSITION 37 Source method: obtained synthetically Details: sequence of tRNA(Lys3) with terminal base pair replaced with GC to enhance synthesis yield and stability |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|---|
| NMR experiment | Type: 2D NOESY |
| NMR details | Text: This structure was determined using standard 2D homonuclear techniques |
- Sample preparation
Sample preparation
| Details | Contents: 1.2 mM RNA, 10 mM cacodylate buffer, pH 5.6, 0.1 mM EDTA Solvent system: 94% H2O, 6% D2O |
|---|---|
| Sample conditions | pH: 5.6 / Pressure: 1 atm / Temperature: 289 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Bruker AVANCE / Manufacturer: Bruker / Model: AVANCE / Field strength: 500 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: global fold by distance geometry. Refinement by simulated annealing using the AMBER forcefield Software ordinal: 1 Details: 272 restraints, 207 NOE-derived, 13 h-bond, 52 dihedral angle. Chi torsion was left unrestrained although likely anti for all residues. | ||||||||||||
| NMR representative | Selection criteria: minimized average structure of family a (struct 2-5) | ||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 50 / Conformers submitted total number: 11 |
 Movie
Movie Controller
Controller








 PDBj
PDBj





























