[English] 日本語
 Yorodumi
Yorodumi- PDB-1aul: SOLUTION STRUCTURE OF A HIGHLY STABLE DNA DUPLEX CONJUGATED TO A ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1aul | ||||||
|---|---|---|---|---|---|---|---|
| Title | SOLUTION STRUCTURE OF A HIGHLY STABLE DNA DUPLEX CONJUGATED TO A MINOR GROOVE BINDER, NMR, MINIMIZED AVERAGE STRUCTURE | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA / DEOXYRIBONUCLEIC ACID / CDPI3 | ||||||
| Function / homology | DNA Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / RESTRAINED MOLECULAR DYNAMICS, RELAXATION MATRIX REFINEMENT | ||||||
 Authors Authors | Kumar, S. / Reed, M.W. / Gamper Junior, H.B. / Gorn, V.V. / Lukhtanov, E.A. / Foti, M. / West, J. / Meyer Junior, R.B. / Schweitzer, B.I. | ||||||
 Citation Citation |  Journal: Nucleic Acids Res. / Year: 1998 Journal: Nucleic Acids Res. / Year: 1998Title: Solution structure of a highly stable DNA duplex conjugated to a minor groove binder. Authors: Kumar, S. / Reed, M.W. / Gamper Jr., H.B. / Gorn, V.V. / Lukhtanov, E.A. / Foti, M. / West, J. / Meyer Jr., R.B. / Schweitzer, B.I. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1aul.cif.gz 1aul.cif.gz | 26.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1aul.ent.gz pdb1aul.ent.gz | 17.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1aul.json.gz 1aul.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1aul_validation.pdf.gz 1aul_validation.pdf.gz | 256.1 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1aul_full_validation.pdf.gz 1aul_full_validation.pdf.gz | 263.5 KB | Display | |
| Data in XML |  1aul_validation.xml.gz 1aul_validation.xml.gz | 4.8 KB | Display | |
| Data in CIF |  1aul_validation.cif.gz 1aul_validation.cif.gz | 5.2 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/au/1aul https://data.pdbj.org/pub/pdb/validation_reports/au/1aul ftp://data.pdbj.org/pub/pdb/validation_reports/au/1aul ftp://data.pdbj.org/pub/pdb/validation_reports/au/1aul | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 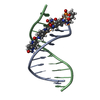
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 3744.794 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: CDPI3 COVALENTLY LINKED TO 5' END OF DNA STRAND A WITH A HEXAMETHYLENE LINKER |
|---|---|
| #2: DNA chain | Mass: 3037.032 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: CDPI3 COVALENTLY LINKED TO 5' END OF DNA STRAND A WITH A HEXAMETHYLENE LINKER |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Sample conditions | pH: 6 / Temperature: 293.15 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Varian UNITYPLUS / Manufacturer: Varian / Model: UNITYPLUS / Field strength: 600 MHz |
|---|
- Processing
Processing
| Software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR software |
| ||||||||||||
| Refinement | Method: RESTRAINED MOLECULAR DYNAMICS, RELAXATION MATRIX REFINEMENT Software ordinal: 1 Details: CANONICAL B-TYPE STRUCTURE USED IN RESTRAINED MOLECULAR DYNAMICS CALCULATIONS GENERATING SIX STRUCTURES FROM DIFFERENT RANDOM NUMBER SEEDS. THE SIX STRUCTURES WERE SUBJECTED TO RELAXATION ...Details: CANONICAL B-TYPE STRUCTURE USED IN RESTRAINED MOLECULAR DYNAMICS CALCULATIONS GENERATING SIX STRUCTURES FROM DIFFERENT RANDOM NUMBER SEEDS. THE SIX STRUCTURES WERE SUBJECTED TO RELAXATION MATRIX REFINEMENT TO GENERATE THE SIX FINAL STRUCTURES. THESE SIX STRUCTURES WERE AVERAGED AND MINIMIZED TO GIVE THE FINAL STRUCTURE PRESENTED HERE. | ||||||||||||
| NMR ensemble | Conformer selection criteria: LEAST RESTRAINT VIOLATION / Conformers calculated total number: 6 / Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller






 PDBj
PDBj

 X-PLOR
X-PLOR