+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1f85 | ||||||
|---|---|---|---|---|---|---|---|
| Title | SOLUTION STRUCTURE OF HCV IRES RNA DOMAIN IIIE | ||||||
 Components Components | HCV-1B IRES RNA DOMAIN IIIE | ||||||
 Keywords Keywords | RNA / Ribonucleic Acid / Hepatitis C virus internal ribosome entry site / Hairpin loop / Tetraloop / RNA structure | ||||||
| Function / homology | RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / Simulated annealing, restrained molecular dynamics | ||||||
 Authors Authors | Lukavsky, P.J. / Otto, G.A. / Lancaster, A.M. / Sarnow, P. / Puglisi, J.D. | ||||||
 Citation Citation |  Journal: Nat.Struct.Biol. / Year: 2000 Journal: Nat.Struct.Biol. / Year: 2000Title: Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Authors: Lukavsky, P.J. / Otto, G.A. / Lancaster, A.M. / Sarnow, P. / Puglisi, J.D. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1f85.cif.gz 1f85.cif.gz | 174.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1f85.ent.gz pdb1f85.ent.gz | 145.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1f85.json.gz 1f85.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/f8/1f85 https://data.pdbj.org/pub/pdb/validation_reports/f8/1f85 ftp://data.pdbj.org/pub/pdb/validation_reports/f8/1f85 ftp://data.pdbj.org/pub/pdb/validation_reports/f8/1f85 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 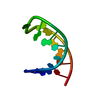
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 4518.731 Da / Num. of mol.: 1 Fragment: HCV IRES RNA DOMAIN IIIE (NUCLEOTIDES 290-303 OF GENOMIC HEPATITIS C VIRAL RNA) Mutation: U290G, G303C / Source method: obtained synthetically Details: Synthesized enzymatically in-vitro using T7 RNA Polymerase |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|---|
| NMR experiment | Type: 31P-decoupled DQF COSY, HP COSY, D2O NOESY, H2O NOESY |
| NMR details | Text: THE STRUCTURE WAS DETERMINED USING A NON-ISOTOPICALLY LABELLED RNA SAMPLE. NATURAL ABUNDANCE 13C HMQC EXPERIMENTS WERE PERFORMED. |
- Sample preparation
Sample preparation
| Details | Contents: 2.5mM RNA, non-isotopically labelled Solvent system: sodium phosphate buffer, 96% H2O/4% D2O or 100% D2O |
|---|---|
| Sample conditions | Ionic strength: 10mM / pH: 6.4 / Pressure: ambient / Temperature: 283 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: Simulated annealing, restrained molecular dynamics / Software ordinal: 1 / Details: SEE REFERENCE ABOVE | ||||||||||||||||||||
| NMR representative | Selection criteria: closest to the average | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the least restraint violations,structures with the lowest energy Conformers calculated total number: 100 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller










 PDBj
PDBj





























 X-PLOR
X-PLOR