[English] 日本語
 Yorodumi
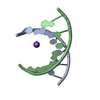
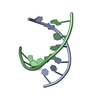
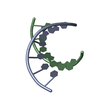
Yorodumi- PDB-2xsl: The crystal structure of a Thermus thermophilus tRNAGly acceptor ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2xsl | ||||||
|---|---|---|---|---|---|---|---|
| Title | The crystal structure of a Thermus thermophilus tRNAGly acceptor stem microhelix at 1.6 Angstroem resolution | ||||||
 Components Components |
| ||||||
 Keywords Keywords | RNA / IDENTITY ELEMENTS / GLYCYL-TRNA SYNTHETASE (GLYRS) / RNA HYDRATION | ||||||
| Function / homology | : / RNA Function and homology information Function and homology information | ||||||
| Biological species |   THERMUS THERMOPHILUS (bacteria) THERMUS THERMOPHILUS (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.59 Å MOLECULAR REPLACEMENT / Resolution: 1.59 Å | ||||||
 Authors Authors | Oberthuer, D. / Eichert, A. / Erdmann, V.A. / Fuerste, J.P. / Betzel, C. / Foerster, C. | ||||||
 Citation Citation |  Journal: Biochem.Biophys.Res.Commun. / Year: 2011 Journal: Biochem.Biophys.Res.Commun. / Year: 2011Title: The Crystal Structure of a Thermus Thermophilus tRNA(Gly) Acceptor Stem Microhelix at 1.6 A Resolution. Authors: Oberthur, D. / Eichert, A. / Erdmann, V.A. / Furste, J.P. / Betzel, C. / Forster, C. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2xsl.cif.gz 2xsl.cif.gz | 28.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2xsl.ent.gz pdb2xsl.ent.gz | 18.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2xsl.json.gz 2xsl.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/xs/2xsl https://data.pdbj.org/pub/pdb/validation_reports/xs/2xsl ftp://data.pdbj.org/pub/pdb/validation_reports/xs/2xsl ftp://data.pdbj.org/pub/pdb/validation_reports/xs/2xsl | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments:
NCS ensembles :
|
- Components
Components
| #1: RNA chain | Mass: 2315.459 Da / Num. of mol.: 2 / Fragment: MICROHELIX, RESIDUES 1-7 / Source method: obtained synthetically / Source: (synth.)   THERMUS THERMOPHILUS (bacteria) / References: GenBank: 55979969 THERMUS THERMOPHILUS (bacteria) / References: GenBank: 55979969#2: RNA chain | Mass: 2132.323 Da / Num. of mol.: 2 / Fragment: MICROHELIX, RESIDUES 66-72 / Source method: obtained synthetically / Source: (synth.)   THERMUS THERMOPHILUS (bacteria) / References: GenBank: 55979969 THERMUS THERMOPHILUS (bacteria) / References: GenBank: 55979969#3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.52 Å3/Da / Density % sol: 62.59 % / Description: NONE |
|---|---|
| Crystal grow | pH: 7 Details: 0.5 MM RNA IN 40 MM SODIUM CACODYLATE, PH 7.0, 12 MM SPERMINE TETRA-HCL, 80 MM NACL AND 10 % (V/V) MPD, EQUILIBRATED AGAINST 35 % (V/V) MPD, ROOM TEMPERATURE. |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  EMBL/DESY, HAMBURG EMBL/DESY, HAMBURG  / Beamline: X13 / Wavelength: 0.8123 / Beamline: X13 / Wavelength: 0.8123 |
| Detector | Type: MARRESEARCH SX-165 / Detector: CCD |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.8123 Å / Relative weight: 1 |
| Reflection | Resolution: 1.6→36 Å / Num. obs: 10142 / % possible obs: 94.7 % / Observed criterion σ(I): 1.7 / Redundancy: 3.4 % / Rmerge(I) obs: 0.07 / Net I/σ(I): 14.5 |
| Reflection shell | Resolution: 1.6→1.63 Å / Redundancy: 2.1 % / Rmerge(I) obs: 0.35 / Mean I/σ(I) obs: 1.7 / % possible all: 72.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: ARTIFICIAL RNA Resolution: 1.59→27.8 Å / Cor.coef. Fo:Fc: 0.952 / Cor.coef. Fo:Fc free: 0.927 / SU B: 3.032 / SU ML: 0.101 / Cross valid method: THROUGHOUT / ESU R: 0.123 / ESU R Free: 0.123 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.59→27.8 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller






 PDBj
PDBj

