+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1.0E+42 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Beta2-adaptin appendage domain, from clathrin adaptor AP2 | ||||||
 Components Components | AP-2 COMPLEX SUBUNIT BETA | ||||||
 Keywords Keywords | ENDOCYTOSIS / ADAPTOR | ||||||
| Function / homology |  Function and homology information Function and homology informationNef Mediated CD8 Down-regulation / WNT5A-dependent internalization of FZD2, FZD5 and ROR2 / Trafficking of GluR2-containing AMPA receptors / clathrin adaptor complex / WNT5A-dependent internalization of FZD4 / extrinsic component of presynaptic endocytic zone membrane / postsynaptic endocytic zone / AP-2 adaptor complex / postsynaptic neurotransmitter receptor internalization / Retrograde neurotrophin signalling ...Nef Mediated CD8 Down-regulation / WNT5A-dependent internalization of FZD2, FZD5 and ROR2 / Trafficking of GluR2-containing AMPA receptors / clathrin adaptor complex / WNT5A-dependent internalization of FZD4 / extrinsic component of presynaptic endocytic zone membrane / postsynaptic endocytic zone / AP-2 adaptor complex / postsynaptic neurotransmitter receptor internalization / Retrograde neurotrophin signalling / clathrin-coated endocytic vesicle / clathrin coat assembly / LDL clearance / clathrin-dependent endocytosis / signal sequence binding / Nef Mediated CD4 Down-regulation / positive regulation of protein localization to membrane / coronary vasculature development / endolysosome membrane / neurotransmitter secretion / ventricular septum development / aorta development / clathrin binding / Recycling pathway of L1 / EPH-ephrin mediated repulsion of cells / positive regulation of endocytosis / synaptic vesicle endocytosis / vesicle-mediated transport / MHC class II antigen presentation / VLDLR internalisation and degradation / intracellular protein transport / kidney development / clathrin-coated endocytic vesicle membrane / cytoplasmic side of plasma membrane / synaptic vesicle / endocytic vesicle membrane / Cargo recognition for clathrin-mediated endocytosis / Clathrin-mediated endocytosis / Potential therapeutics for SARS / protein-containing complex binding / glutamatergic synapse / membrane / plasma membrane / cytosol Similarity search - Function | ||||||
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  SIRAS / Resolution: 1.7 Å SIRAS / Resolution: 1.7 Å | ||||||
 Authors Authors | Owen, D.J. / Evans, P.R. / McMahon, H.T. | ||||||
 Citation Citation |  Journal: Embo J. / Year: 2000 Journal: Embo J. / Year: 2000Title: The Structure and Function of the Beta2-Adaptin Appendage Domain Authors: Owen, D.J. / Vallis, Y. / Pearse, B.M.F. / Mcmahon, H.T. / Evans, P.R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1e42.cif.gz 1e42.cif.gz | 128 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1e42.ent.gz pdb1e42.ent.gz | 98.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1e42.json.gz 1e42.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/e4/1e42 https://data.pdbj.org/pub/pdb/validation_reports/e4/1e42 ftp://data.pdbj.org/pub/pdb/validation_reports/e4/1e42 ftp://data.pdbj.org/pub/pdb/validation_reports/e4/1e42 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Noncrystallographic symmetry (NCS) | NCS oper: (Code: given Matrix: (-0.379985, 0.020033, -0.924776), Vector: Details | THE BETA-ADAPTIN PROTEIN IS PART OF THE ASSEMBLY PROTEINCOMPLEX 2, (AP-2), THAT IS A HETEROTETRAMER COMPOSED OF TWO LARGE CHAINS (ALPHA AND BETA), A MEDIUM CHAIN (AP50)AND A SMALL CHAIN (AP17). | |
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 29088.453 Da / Num. of mol.: 2 / Fragment: APPENDAGE DOMAIN, RESIDUES 701-937 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Plasmid: PET15B / Production host: HOMO SAPIENS (human) / Plasmid: PET15B / Production host:  |
|---|
-Non-polymers , 6 types, 835 molecules 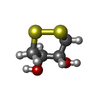










| #2: Chemical | | #3: Chemical | ChemComp-MG / #4: Chemical | ChemComp-CL / #5: Chemical | #6: Chemical | ChemComp-NI / | #7: Water | ChemComp-HOH / | |
|---|
-Details
| Sequence details | CLONING HEADER MGSSHHHHHH |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.9 Å3/Da / Density % sol: 55 % | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 8.8 Details: WELL SOLUTION: 1.6 - 2.0M MGCL2, 0.1M BICINE PH 8.7 - 9.0, 15% GLYCEROL, 1MM DTT. PROTEIN: 40MG/ML, 5MM HEPES, 50MM NACL, 4MM DTT, 1:1 MIXTURE | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop / PH range low: 9 / PH range high: 8.7 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SRS SRS  / Beamline: PX9.6 / Wavelength: 0.88 / Beamline: PX9.6 / Wavelength: 0.88 |
| Detector | Type: ADSC QUANTUM / Detector: CCD / Date: Sep 15, 1998 |
| Radiation | Monochromator: SI(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.88 Å / Relative weight: 1 |
| Reflection | Resolution: 1.7→19.96 Å / Num. obs: 71562 / % possible obs: 98.4 % / Observed criterion σ(I): 4 / Redundancy: 6.85 % / Biso Wilson estimate: 33 Å2 / Rmerge(I) obs: 0.157 / Rsym value: 0.157 / Net I/σ(I): 11.2 |
| Reflection shell | Resolution: 1.7→1.79 Å / Redundancy: 4.8 % / Rmerge(I) obs: 0.75 / Mean I/σ(I) obs: 1.48 / Rsym value: 0.75 / % possible all: 89.6 |
| Reflection shell | *PLUS % possible obs: 98.3 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  SIRAS / Resolution: 1.7→67.42 Å / SU B: 3.538 / SU ML: 0.116 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.179 / ESU R Free: 0.164 SIRAS / Resolution: 1.7→67.42 Å / SU B: 3.538 / SU ML: 0.116 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.179 / ESU R Free: 0.164 Details: RESIDUES A980 AND B980 WERE MODELLED AS GLYCEROL, BUT IDENTIFICATION IS NOT CERTAIN. RESIDUE B970 WAS MODELLED AS A 50% NI ION, SINCE IT IS BOUND TO A HISTIDINE, BUT IT MAY BE ANOTHER MG ION
| ||||||||||||||||||||
| Displacement parameters | Biso mean: 32 Å2
| ||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.7→67.42 Å
| ||||||||||||||||||||
| Software | *PLUS Name: REFMAC / Version: VERSION 5 / Classification: refinement | ||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.196 | ||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller











 PDBj
PDBj






















