+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1e3d | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | [NiFe] Hydrogenase from Desulfovibrio desulfuricans ATCC 27774 | |||||||||
 Components Components | ([NiFe] hydrogenase ...) x 2 | |||||||||
 Keywords Keywords | HYDROGENASE / MOLECULAR MODELLING / ELECTRON TRANSFER | |||||||||
| Function / homology |  Function and homology information Function and homology informationcytochrome-c3 hydrogenase / cytochrome-c3 hydrogenase activity / ferredoxin hydrogenase complex / [Ni-Fe] hydrogenase complex / ferredoxin hydrogenase activity / anaerobic respiration / 3 iron, 4 sulfur cluster binding / nickel cation binding / 4 iron, 4 sulfur cluster binding / electron transfer activity ...cytochrome-c3 hydrogenase / cytochrome-c3 hydrogenase activity / ferredoxin hydrogenase complex / [Ni-Fe] hydrogenase complex / ferredoxin hydrogenase activity / anaerobic respiration / 3 iron, 4 sulfur cluster binding / nickel cation binding / 4 iron, 4 sulfur cluster binding / electron transfer activity / periplasmic space / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Desulfovibrio desulfuricans (bacteria) Desulfovibrio desulfuricans (bacteria) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.8 Å MOLECULAR REPLACEMENT / Resolution: 1.8 Å | |||||||||
 Authors Authors | Matias, P.M. / Soares, C.M. / Saraiva, L.M. / Coelho, R. / Morais, J. / Le Gall, J. / Carrondo, M.A. | |||||||||
 Citation Citation |  Journal: J.Biol.Inorg.Chem. / Year: 2001 Journal: J.Biol.Inorg.Chem. / Year: 2001Title: [Nife] Hydrogenase from Desulfovibrio Desulfuricans Atcc 27774: Gene Sequencing, Three-Dimensional Structure Determination and Refinement at 1.8 A and Modelling Studies of its Interaction with ...Title: [Nife] Hydrogenase from Desulfovibrio Desulfuricans Atcc 27774: Gene Sequencing, Three-Dimensional Structure Determination and Refinement at 1.8 A and Modelling Studies of its Interaction with the Tetrahaem Cytochrome C3. Authors: Matias, P.M. / Soares, C.M. / Saraiva, L.M. / Coelho, R. / Morais, J. / Le Gall, J. / Carrondo, M.A. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1e3d.cif.gz 1e3d.cif.gz | 606.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1e3d.ent.gz pdb1e3d.ent.gz | 502.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1e3d.json.gz 1e3d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/e3/1e3d https://data.pdbj.org/pub/pdb/validation_reports/e3/1e3d ftp://data.pdbj.org/pub/pdb/validation_reports/e3/1e3d ftp://data.pdbj.org/pub/pdb/validation_reports/e3/1e3d | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1frvS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| 2 | 
| ||||||||||||
| Unit cell |
| ||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
|
- Components
Components
-[NiFe] hydrogenase ... , 2 types, 4 molecules ACBD
| #1: Protein | Mass: 28682.602 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Desulfovibrio desulfuricans (bacteria) / References: UniProt: Q9L869 Desulfovibrio desulfuricans (bacteria) / References: UniProt: Q9L869#2: Protein | Mass: 60009.902 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Desulfovibrio desulfuricans (bacteria) / References: UniProt: Q9L868 Desulfovibrio desulfuricans (bacteria) / References: UniProt: Q9L868 |
|---|
-Non-polymers , 7 types, 1413 molecules 


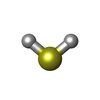
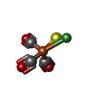




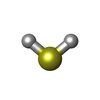
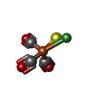


| #3: Chemical | | #4: Chemical | #5: Chemical | #6: Chemical | ChemComp-H2S / #7: Chemical | #8: Chemical | #9: Water | ChemComp-HOH / | |
|---|
-Details
| Has protein modification | Y |
|---|---|
| Sequence details | THERE IS AN INITIAL MET 1 THAT WAS NOT CONSIDERED IN THE SEQUENCE USED FOR THE CRYSTALLOGRAPHIC ...THERE IS AN INITIAL MET 1 THAT WAS NOT CONSIDERED |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.23 Å3/Da / Density % sol: 44.78 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 8.5 Details: 25% (W/V) PEG 4000, 0.1 M TRIS-HCL BUFFER PH 8.5, 0.2 M MGCL2 AND A TRACE AMOUNT OF A DETERGENT, C12-DAO. | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, sitting drop | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-4 / Wavelength: 0.932 / Beamline: ID14-4 / Wavelength: 0.932 |
| Detector | Type: ADSC CCD / Detector: CCD / Date: May 15, 1999 / Details: TOROIDAL MIRROR |
| Radiation | Monochromator: DOUBLE CRYSTAL SI 111 / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.932 Å / Relative weight: 1 |
| Reflection | Resolution: 1.8→32 Å / Num. obs: 135264 / % possible obs: 95.4 % / Redundancy: 2.5 % / Rmerge(I) obs: 0.072 / Net I/σ(I): 4.5 |
| Reflection shell | Resolution: 1.8→1.9 Å / Redundancy: 2.3 % / Rmerge(I) obs: 0.412 / Mean I/σ(I) obs: 1.7 / % possible all: 96 |
| Reflection | *PLUS Num. measured all: 337524 |
| Reflection shell | *PLUS % possible obs: 96 % |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1FRV Resolution: 1.8→25 Å / Num. parameters: 55536 / Num. restraintsaints: 52359 / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: ENGH AND HUBER Details: FOR EACH CHAIN THE N-TERMINAL RESIDUES 1-4 (CHAINS A AND C) AND 1-5 (CHAINS B AND D) WERE NOT OBSERVED IN THE DENSITY MAPS
| |||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: MOEWS & KRETSINGER | |||||||||||||||||||||||||||||||||
| Refine analyze | Num. disordered residues: 6 / Occupancy sum hydrogen: 11896.01 / Occupancy sum non hydrogen: 13663.51 | |||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.8→25 Å
| |||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||
| Software | *PLUS Name: SHELXL-97 / Classification: refinement | |||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor Rwork: 0.1671 | |||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller























 PDBj
PDBj









