+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1dy2 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Murine collagen alpha1(XV), endostatin domain | ||||||
 Components Components | COLLAGEN ALPHA1(XV) CHAIN | ||||||
 Keywords Keywords | ANGIOGENESIS INHIBITOR | ||||||
| Function / homology |  Function and homology information Function and homology informationCollagen biosynthesis and modifying enzymes / Collagen chain trimerization / Collagen degradation / Assembly of collagen fibrils and other multimeric structures / extracellular matrix structural constituent conferring tensile strength / collagen trimer / basement membrane / extracellular matrix / cell adhesion / endoplasmic reticulum / extracellular space Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å | ||||||
 Authors Authors | Tisi, D. / Hohenester, E. / Sasaki, T. / Timpl, R. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2000 Journal: J.Mol.Biol. / Year: 2000Title: Endostatin Derived from Collagens Xv and Xviii Differ in Structural and Binding Properties, Tissue Distribution and Anti-Angiogenic Activity Authors: Sasaki, T. / Larsson, H. / Tisi, D. / Claesson-Welsh, L. / Hohenester, E. / Timpl, R. #1:  Journal: Embo J. / Year: 1998 Journal: Embo J. / Year: 1998Title: Crystal Structure of the Angiogenesis Inhibitor Endostatin at 1.5 Angstrom Resolution Authors: Hohenester, E. / Sasaki, T. / Olsen, B.R. / Timpl, R. #2: Journal: Cell(Cambridge,Mass.) / Year: 1997 Title: Endostatin: An Endogenous Inhibitor of Angiogenesis and Tumor Growth Authors: O'Reilly, M.S. / Boehm, T. / Shing, Y. / Fukai, N. / Vasios, G. / Lane, W.S. / Flynn, E. / Birkhead, J.R. / Olsen, B.R. / Folkman, J. | ||||||
| History |
|
- Structure visualization
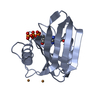
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1dy2.cif.gz 1dy2.cif.gz | 46.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1dy2.ent.gz pdb1dy2.ent.gz | 32.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1dy2.json.gz 1dy2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dy/1dy2 https://data.pdbj.org/pub/pdb/validation_reports/dy/1dy2 ftp://data.pdbj.org/pub/pdb/validation_reports/dy/1dy2 ftp://data.pdbj.org/pub/pdb/validation_reports/dy/1dy2 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1koeS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Details | BIOLOGICAL UNIT IS A MONOMER |
- Components
Components
| #1: Protein | Mass: 20085.875 Da / Num. of mol.: 1 / Fragment: ENDOSTATIN DOMAIN Source method: isolated from a genetically manipulated source Source: (gene. exp.)   HOMO SAPIENS (human) / References: UniProt: O35206 HOMO SAPIENS (human) / References: UniProt: O35206 |
|---|---|
| #2: Chemical | ChemComp-SO4 / |
| #3: Water | ChemComp-HOH / |
| Has protein modification | Y |
| Sequence details | N-TERMINAL APLA |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal grow | pH: 6.5 / Details: pH 6.50 | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS pH: 8 / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 293 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Wavelength: 1.5418 ROTATING ANODE / Wavelength: 1.5418 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Mar 15, 1999 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2→20 Å / Num. obs: 9225 / % possible obs: 97.1 % / Redundancy: 4.7 % / Rmerge(I) obs: 0.069 |
| Reflection shell | Resolution: 2→2.12 Å / Redundancy: 4.7 % / Rmerge(I) obs: 0.152 / % possible all: 97.2 |
| Reflection | *PLUS Rmerge(I) obs: 0.06 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1KOE Resolution: 2→20 Å / Isotropic thermal model: INDIVIDUAL RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Classification: refinement X-PLOR / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.2 / Rfactor Rwork: 0.2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller














 PDBj
PDBj









