+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1dxa | ||||||
|---|---|---|---|---|---|---|---|
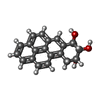
| Title | BENZO[A]PYRENE DIOL EPOXIDE ADDUCT OF DA IN DUPLEX DNA | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA / DOUBLE HELIX / BENZO[A]PYRENE DIOL EPOXIDE ADDUCT / DUPLEX DNA / NON-WATSON-CRICK BASE PAIR | ||||||
| Function / homology | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE / DNA Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / ENERGY MINIMIZATION, MOLECULAR DYNAMICS | ||||||
 Authors Authors | Yeh, H.J.C. / Sayer, J.M. / Liu, X. / Altieri, A.S. / Byrd, R.A. / Lakshman, M.K. / Yagi, H. / Schurter, E.J. / Gorenstein, D.G. / Jerina, D.M. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1995 Journal: Biochemistry / Year: 1995Title: NMR solution structure of a nonanucleotide duplex with a dG mismatch opposite a 10S adduct derived from trans addition of a deoxyadenosine N6-amino group to (+)-(7R,8S,9S,10R)-7,8-dihydroxy- ...Title: NMR solution structure of a nonanucleotide duplex with a dG mismatch opposite a 10S adduct derived from trans addition of a deoxyadenosine N6-amino group to (+)-(7R,8S,9S,10R)-7,8-dihydroxy-9,10-epoxy-7,8,9,10- tetrahydrobenzo[a]pyrene: an unusual syn glycosidic torsion angle at the modified dA Authors: Yeh, H.J. / Sayer, J.M. / Liu, X. / Altieri, A.S. / Byrd, R.A. / Lakshman, M.K. / Yagi, H. / Schurter, E.J. / Gorenstein, D.G. / Jerina, D.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1dxa.cif.gz 1dxa.cif.gz | 22.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1dxa.ent.gz pdb1dxa.ent.gz | 14 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1dxa.json.gz 1dxa.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dx/1dxa https://data.pdbj.org/pub/pdb/validation_reports/dx/1dxa ftp://data.pdbj.org/pub/pdb/validation_reports/dx/1dxa ftp://data.pdbj.org/pub/pdb/validation_reports/dx/1dxa | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 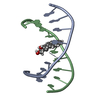
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 2780.836 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: chemically synthesized /  Keywords: DEOXYRIBONUCLEIC ACID Keywords: DEOXYRIBONUCLEIC ACID |
|---|---|
| #2: DNA chain | Mass: 2716.787 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: chemically synthesized /  Keywords: DEOXYRIBONUCLEIC ACID Keywords: DEOXYRIBONUCLEIC ACID |
| #3: Chemical | ChemComp-BAP / |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|---|
| NMR details | Text: (+)-(7R,8S,9S,10R)-7,8-DIHYDROXY-9,10-EPOXY-7,8,9,10- TETRAHYDROBENZO[A]PYRENE IS COVALENTLY BONDED TO THE EXOCYCLIC N6 AMINO GROUP OF DEOXYADENOSINE IN THE CENTER OF THE DUPLEX THROUGH TRANS ...Text: (+)-(7R,8S,9S,10R)-7,8-DIHYDROXY-9,10-EPOXY-7,8,9,10- TETRAHYDROBENZO[A]PYRENE IS COVALENTLY BONDED TO THE EXOCYCLIC N6 AMINO GROUP OF DEOXYADENOSINE IN THE CENTER OF THE DUPLEX THROUGH TRANS ADDITION AT THE C10 OF THE EPOXIDE. |
- Sample preparation
Sample preparation
| Crystal grow | *PLUS Method: other |
|---|
- Processing
Processing
| NMR software | Name: NMRCHITECT / Developer: BIOSYM / Classification: refinement |
|---|---|
| Refinement | Method: ENERGY MINIMIZATION, MOLECULAR DYNAMICS / Software ordinal: 1 Details: TWO STARTING STRUCTURES WITH BENZO[A]PYRENE MOIETY INSIDE THE DUPLEX AND OUTSIDE THE DUPLEX, RESPECTIVELY, WERE GENERATED BY CONNECTING A STANDARD B-TYPE DNA FRAGMENT WITH A MANUALLY BUILD ...Details: TWO STARTING STRUCTURES WITH BENZO[A]PYRENE MOIETY INSIDE THE DUPLEX AND OUTSIDE THE DUPLEX, RESPECTIVELY, WERE GENERATED BY CONNECTING A STANDARD B-TYPE DNA FRAGMENT WITH A MANUALLY BUILD (+)-DE2-[BAP] FRAGMENT. SOLUTION STRUCTURES FOR THE DUPLEX WERE GENERATED BY RESTRAINTED MOLECULAR DYNAMICS AND RESTRAINTED ENERGY MINIMIZATION USING A SET OF INTER-PROTON DISTANCE RESTRAINTS AND DIHEDRAL ANGLE RESTRAINTS WHICH WERE DERIVED FROM NMR EXPERIMENTS. SOME OF STRUCTURES WITH MINIMUM RESTRAINT VIOLATIONS WERE FURTHER REFINED BY THE ITERATIVE RELAXATION MATRIX ANALYSIS (IRMA) METHOD OVER 111 WELL-SEPARATED CROSS PEAKS IN NOESY SPECTRA RECORDED IN D2O AND MIXING TIMES OF 50, 80, 120, 160 AND 200 MS. THE STRUCTURE PRESENTED HERE WAS ONE OF THE FIVE REFINED AND MINIMIZED STRUCTURES. THE FOLLOWING RESTRAINTS WERE APPLIED IN THE MOLECULAR DYNAMICS/ENERGY MINIMIZATION CALCULATIONS. THESE WERE; (1) INTER-PROTON DISTANCE RESTRAINTS DIRECTLY DERIVED FROM NOE DATA; (2) BACKBONE DIHEDRAL ANGLE RESTRAINTS FOR A STANDARD B-DNA EXCEPT THOSE IMMEDIATELY ADJACENT TO THE MODIFIED RESIDUE; (3) DIHEDRAL ANGLE RESTRAINTS FOR ALL GLYCOSIDIC BONDS, EXCEPT MODIFIED DA RESIDUE IN THE CENTER; (4) HYDROGEN BOND RESTRAINTS (BOTH DISTANCE AND PLANARITY ) FOR ALL BASE-PAIRS EXCEPT CENTRAL DA-DG MISMATCH PAIR. THESE RESTRAINTS WERE JUSTIFIED BY THE FACT THAT THE MODIFIED DUPLEX DISPLAYED CHARACTERISTIC NMR SPECTRA OF A B-DNA EXCEPT NEAR LESION SITE, WHERE THE BAP MOIETY WAS INTERCALATED. THE MODIFIED DA RESIDUE DISPLAYED C3'-ENDO SUGAR RING AND A SYN GLYCOSIDIC BOND. IT ALSO FORMED A NON-WATSON-CRICK BASE PAIR WITH THE OPPOSITE DG. |
| NMR ensemble | Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller





 PDBj
PDBj