[English] 日本語
 Yorodumi
Yorodumi- PDB-1c2q: SOLUTION STRUCTURE OF A DNA.RNA HYBRID CONTAINING AN ALPHAT-ANOME... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1c2q | ||||||
|---|---|---|---|---|---|---|---|
| Title | SOLUTION STRUCTURE OF A DNA.RNA HYBRID CONTAINING AN ALPHAT-ANOMERIC THYMIDINE AND POLARITY REVERSALS | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA-RNA HYBRID / DNA/RNA HYBRID / ALPHAT-ANOMERIC THYMIDINE / 3'-3'/5'-5' PHOSPHODIESTER LINKAGES | ||||||
| Function / homology | DNA / RNA Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / SIMULATED ANNEALING, MOLECULAR DYNAMICS | ||||||
| Model type details | minimized average | ||||||
 Authors Authors | Germann, M.W. / Aramini, J.M. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1999 Journal: Biochemistry / Year: 1999Title: Solution structure of a DNA.RNA hybrid containing an alpha-anomeric thymidine and polarity reversals: d(ATGG-3'-3'-alphaT-5'-5'-GCTC). r(gagcaccau). Authors: Aramini, J.M. / Germann, M.W. #1:  Journal: Nucleic Acids Res. / Year: 1998 Journal: Nucleic Acids Res. / Year: 1998Title: NMR solution structures of [d(GCGAAT-3'-3'-(alpha-T)-5'-5'-CGC)2] and its unmodified control Authors: Aramini, J.M. / Mujeeb, A. / Germann, M.W. | ||||||
| History |
|
- Structure visualization
Structure visualization
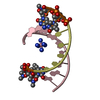
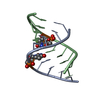
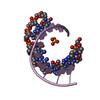
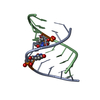
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1c2q.cif.gz 1c2q.cif.gz | 23.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1c2q.ent.gz pdb1c2q.ent.gz | 13.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1c2q.json.gz 1c2q.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/c2/1c2q https://data.pdbj.org/pub/pdb/validation_reports/c2/1c2q ftp://data.pdbj.org/pub/pdb/validation_reports/c2/1c2q ftp://data.pdbj.org/pub/pdb/validation_reports/c2/1c2q | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 2746.809 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: SYNTHETIC |
|---|---|
| #2: RNA chain | Mass: 2854.782 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: SYNTHETIC |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||
| NMR details | Text: THIS STRUCTURE WAS DETERMINED USING STANDARD 2D HOMONUCLEAR NMR TECHNIQUES |
- Sample preparation
Sample preparation
| Details |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions |
| ||||||||||||
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Bruker AMX / Manufacturer: Bruker / Model: AMX / Field strength: 600 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: SIMULATED ANNEALING, MOLECULAR DYNAMICS / Software ordinal: 1 Details: THE STRUCTURE IS BASED ON A TOTAL OF 405 RESTRAINTS: 268 RANDMARDI DERIVED INTERPROTON DISTANCE RESTRAINTS (NON-EXCHANGEABLE: 246; EXCHANGEABLE: 22); 45 DEOXYRIBOSE ENDOCYCLIC TORSION ANGLE ...Details: THE STRUCTURE IS BASED ON A TOTAL OF 405 RESTRAINTS: 268 RANDMARDI DERIVED INTERPROTON DISTANCE RESTRAINTS (NON-EXCHANGEABLE: 246; EXCHANGEABLE: 22); 45 DEOXYRIBOSE ENDOCYCLIC TORSION ANGLE RESTRAINTS DERIVED FROM PSEUDOROTATION ANALYSIS; 45 RIBOSE ENDOCYCLIC TORSION ANGLE RESTRAINTS (BROAD, N-TYPE); 46 WATSON-CRICK DISTANCE AND ANGLE RESTRAINTS. THE FINAL AVERAGE STRUCTURE WAS OBTAINED BY COORDINATE AVERAGING OF THE FINAL ENSEMBLE OF RMD/REM STRUCTURES, FOLLOWED BY RESTRAINED ENERGY MINIMIZATION. ALL STRUCTURE CALCULATIONS WERE PERFORMED USING THE SANDER PROGRAM WITHIN AMBER 4.1, AND THE 1994 ALL ATOM NUCLEIC ACID PARAMETERIZATION. ALL CALCULATIONS WERE CONDUCTED IN VACUO, USING A DISTANCE DEPENDENT DIELECTRIC CONSTANT AND 30 A CUT-OFF FOR NON-BONDED INTERACTIONS. | ||||||||||||||||||||||||||||||||||||
| NMR representative | Selection criteria: minimized average structure | ||||||||||||||||||||||||||||||||||||
| NMR ensemble | Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller




 PDBj
PDBj Amber
Amber