[English] 日本語
 Yorodumi
Yorodumi- PDB-1bcx: MUTATIONAL AND CRYSTALLOGRAPHIC ANALYSES OF THE ACTIVE SITE RESID... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1bcx | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | MUTATIONAL AND CRYSTALLOGRAPHIC ANALYSES OF THE ACTIVE SITE RESIDUES OF THE BACILLUS CIRCULANS XYLANASE | |||||||||
 Components Components | XYLANASE | |||||||||
 Keywords Keywords | HYDROLASE(XYLAN DEGRADATION) | |||||||||
| Function / homology |  Function and homology information Function and homology informationendo-1,4-beta-xylanase activity / endo-1,4-beta-xylanase / xylan catabolic process Similarity search - Function | |||||||||
| Biological species |  Bacillus circulans (bacteria) Bacillus circulans (bacteria) | |||||||||
| Method |  X-RAY DIFFRACTION / Resolution: 1.81 Å X-RAY DIFFRACTION / Resolution: 1.81 Å | |||||||||
 Authors Authors | Campbell, R.L. / Wakarchuk, W.W. | |||||||||
 Citation Citation |  Journal: Protein Sci. / Year: 1994 Journal: Protein Sci. / Year: 1994Title: Mutational and crystallographic analyses of the active site residues of the Bacillus circulans xylanase. Authors: Wakarchuk, W.W. / Campbell, R.L. / Sung, W.L. / Davoodi, J. / Yaguchi, M. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1bcx.cif.gz 1bcx.cif.gz | 51.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1bcx.ent.gz pdb1bcx.ent.gz | 36 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1bcx.json.gz 1bcx.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1bcx_validation.pdf.gz 1bcx_validation.pdf.gz | 431.9 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1bcx_full_validation.pdf.gz 1bcx_full_validation.pdf.gz | 431.8 KB | Display | |
| Data in XML |  1bcx_validation.xml.gz 1bcx_validation.xml.gz | 5.2 KB | Display | |
| Data in CIF |  1bcx_validation.cif.gz 1bcx_validation.cif.gz | 8.1 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bc/1bcx https://data.pdbj.org/pub/pdb/validation_reports/bc/1bcx ftp://data.pdbj.org/pub/pdb/validation_reports/bc/1bcx ftp://data.pdbj.org/pub/pdb/validation_reports/bc/1bcx | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Atom site foot note | 1: CIS PROLINE - PRO 75 2: SITE AS1 (GLU 78, CYS 172) REFERS TO THE CATALYTIC ACIDIC RESIDUES. IN THE NATIVE SEQUENCE THEY ARE GLU 78 AND GLU 172. SITE AS2 (TYR 5, TYR 69, TYR 80, TYR 166) REFERS TO THE GROUP OF TYROSINE ...2: SITE AS1 (GLU 78, CYS 172) REFERS TO THE CATALYTIC ACIDIC RESIDUES. IN THE NATIVE SEQUENCE THEY ARE GLU 78 AND GLU 172. SITE AS2 (TYR 5, TYR 69, TYR 80, TYR 166) REFERS TO THE GROUP OF TYROSINE RESIDUES IN THE ACTIVE SITE THAT APPEAR TO BE IMPORTANT FOR SUBSTRATE BINDING. 3: ALL NON-GLYCINE RESIDUES LIE WITHIN THE ALLOWED REGIONS OF THE RAMACHANDRAN PLOT EXCEPT ASP 121 AND ALA 165. THE ELECTRON DENSITY FOR BOTH ASP 121 AND ALA 165 IS VERY CLEAR. |
- Components
Components
| #1: Protein | Mass: 20383.033 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Bacillus circulans (bacteria) / References: UniProt: P09850, endo-1,4-beta-xylanase Bacillus circulans (bacteria) / References: UniProt: P09850, endo-1,4-beta-xylanase |
|---|---|
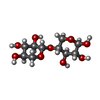
| #2: Polysaccharide | beta-D-xylopyranose-(1-4)-beta-D-xylopyranose / 4beta-beta-xylobiose |
| #3: Chemical | ChemComp-SO4 / |
| #4: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.25 Å3/Da / Density % sol: 45.34 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop / Details: used to seed | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Radiation | Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| Reflection | *PLUS Highest resolution: 1.49 Å / Lowest resolution: 8 Å / Rmerge(I) obs: 0.165 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 1.81→8 Å / σ(F): 2 Details: ALL NON-GLYCINE RESIDUES LIE WITHIN THE ALLOWED REGIONS OF THE RAMACHANDRAN PLOT EXCEPT ASP 121 AND ALA 165. THE ELECTRON DENSITY FOR BOTH ASP 121 AND ALA 165 IS VERY CLEAR.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.81→8 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Classification: refinement X-PLOR / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.161 / Rfactor Rwork: 0.161 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS Type: x_angle_d / Dev ideal: 1.66 |
 Movie
Movie Controller
Controller











 PDBj
PDBj