+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1b6f | ||||||
|---|---|---|---|---|---|---|---|
| Title | BIRCH POLLEN ALLERGEN BET V 1 | ||||||
 Components Components | PROTEIN (MAJOR POLLEN ALLERGEN BET V 1-A) | ||||||
 Keywords Keywords | PLANT PROTEIN / MAJOR BIRCH POLLEN ALLERGEN / PATHOGENESIS-RELATED PROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology informationabscisic acid binding / abscisic acid-activated signaling pathway / protein phosphatase inhibitor activity / defense response / signaling receptor activity / nucleus / cytoplasm Similarity search - Function | ||||||
| Biological species |  Betula pendula (European white birch) Betula pendula (European white birch) | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
 Authors Authors | Schweimer, K. / Sticht, H. / Boehm, M. / Roesch, P. | ||||||
 Citation Citation | Journal: APPL.MAGN.RESON. / Year: 1999 Title: NMR Spectroscopy Reveals Common Structural Features of the Birch Pollen Allergen Bet v 1 and the cherry allergen Pru a 1 Authors: Schweimer, K. / Sticht, H. / Boehm, M. / Roesch, P. #1:  Journal: Biol.Chem. / Year: 1997 Journal: Biol.Chem. / Year: 1997Title: Expression in Escherichia Coli, Purification, and Spectroscopic Characterization of Two Mutant Bet V 1 Proteins Authors: Boehm, M. / Roesch, P. #2:  Journal: J.Biol.Chem. / Year: 1996 Journal: J.Biol.Chem. / Year: 1996Title: Secondary Structure and Tertiary Fold of the Birch Pollen Allergen Bet V 1 in Solution Authors: Faber, C. / Lindemann, A. / Sticht, H. / Ejchart, A. / Kungl, A. / Susani, M. / Frank, R.W. / Kraft, D. / Breitenbach, M. / Roesch, P. | ||||||
| History |
|
- Structure visualization
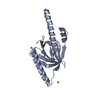
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1b6f.cif.gz 1b6f.cif.gz | 1.1 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1b6f.ent.gz pdb1b6f.ent.gz | 930.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1b6f.json.gz 1b6f.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/b6/1b6f https://data.pdbj.org/pub/pdb/validation_reports/b6/1b6f ftp://data.pdbj.org/pub/pdb/validation_reports/b6/1b6f ftp://data.pdbj.org/pub/pdb/validation_reports/b6/1b6f | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 17443.557 Da / Num. of mol.: 1 / Mutation: M139L Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Betula pendula (European white birch) / Description: SYNTHETIC GENE / Cell: POLLEN / Species (production host): Escherichia coli / Production host: Betula pendula (European white birch) / Description: SYNTHETIC GENE / Cell: POLLEN / Species (production host): Escherichia coli / Production host:  |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||
| NMR details | Text: THE STRUCTURE WAS DETERMINED USING MULTIDIMENSIONAL HETERONUCLEAR NMR SPECTROSCOPY ON 13C, 15N-LABELED BET V 1. |
- Sample preparation
Sample preparation
| Sample conditions | Ionic strength: 10 mM / pH: 7 / Pressure: 1 atm / Temperature: 298 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Bruker DRX600 / Manufacturer: Bruker / Model: DRX600 / Field strength: 600 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 Details: REFINEMENT DETAILS CAN BE FOUND IN THE JRNL CITATION ABOVE | ||||||||||||||||
| NMR ensemble | Conformer selection criteria: LOWEST ENERGY AND LEAST RESTRAINT VIOLATION Conformers calculated total number: 60 / Conformers submitted total number: 23 |
 Movie
Movie Controller
Controller












 PDBj
PDBj HNCA
HNCA