[English] 日本語
 Yorodumi
Yorodumi- PDB-1b2i: KRINGLE 2 DOMAIN OF HUMAN PLASMINOGEN: NMR SOLUTION STRUCTURE OF ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1b2i | ||||||
|---|---|---|---|---|---|---|---|
| Title | KRINGLE 2 DOMAIN OF HUMAN PLASMINOGEN: NMR SOLUTION STRUCTURE OF TRANS-4-AMINOMETHYLCYCLOHEXANE-1-CARBOXYLIC ACID (AMCHA) COMPLEX | ||||||
 Components Components | PROTEIN (PLASMINOGEN) | ||||||
 Keywords Keywords | HYDROLASE / SERINE PROTEASE / FIBRINOLYSIS / LYSINE-BINDING DOMAIN / PLASMINOGEN / KRINGLE 2 | ||||||
| Function / homology |  Function and homology information Function and homology informationplasmin / tissue remodeling / protein antigen binding / Signaling by PDGF / positive regulation of fibrinolysis / negative regulation of cell-cell adhesion mediated by cadherin / Dissolution of Fibrin Clot / biological process involved in interaction with symbiont / Activation of Matrix Metalloproteinases / apolipoprotein binding ...plasmin / tissue remodeling / protein antigen binding / Signaling by PDGF / positive regulation of fibrinolysis / negative regulation of cell-cell adhesion mediated by cadherin / Dissolution of Fibrin Clot / biological process involved in interaction with symbiont / Activation of Matrix Metalloproteinases / apolipoprotein binding / extracellular matrix disassembly / positive regulation of blood vessel endothelial cell migration / negative regulation of cell-substrate adhesion / negative regulation of fibrinolysis / fibrinolysis / Degradation of the extracellular matrix / serine-type peptidase activity / platelet alpha granule lumen / kinase binding / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / blood coagulation / Platelet degranulation / protein-folding chaperone binding / protease binding / : / endopeptidase activity / blood microparticle / protein domain specific binding / external side of plasma membrane / signaling receptor binding / negative regulation of cell population proliferation / serine-type endopeptidase activity / enzyme binding / cell surface / proteolysis / extracellular space / extracellular exosome / extracellular region / plasma membrane Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | SOLUTION NMR / DISTANCE GEOMETRY, DYNAMIC SIMULATED ANNEALING | ||||||
 Authors Authors | Marti, D.N. / Schaller, J. / Llinas, M. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1999 Journal: Biochemistry / Year: 1999Title: Solution structure and dynamics of the plasminogen kringle 2-AMCHA complex: 3(1)-helix in homologous domains. Authors: Marti, D.N. / Schaller, J. / Llinas, M. #1:  Journal: Biochemistry / Year: 1997 Journal: Biochemistry / Year: 1997Title: Ligand Preferences of Kringle 2 and Homologous Domains of Human Plasminogen: Canvassing Weak, Intermediate and High-Affinity Binding Sites by 1H-NMR Authors: Marti, D.N. / Hu, C.K. / An, S.S.A. / Von Haller, P. / Schaller, J. / Llinas, M. #2:  Journal: Biochemistry / Year: 1996 Journal: Biochemistry / Year: 1996Title: Recombinant Gene Expression and 1H NMR Characteristics of the Kringle (2+3) Supermodule: Spectroscopic/Functional Individuality of Plasminogen Kringle Domains Authors: Soehndel, S. / Hu, C.K. / Marti, D. / Affolter, M. / Schaller, J. / Llinas, M. / Rickli, E.E. #3:  Journal: Eur.J.Biochem. / Year: 1994 Journal: Eur.J.Biochem. / Year: 1994Title: Expression, Purification and Characterization of the Recombinant Kringle 2 and Kringle 3 Domains of Human Plasminogen and Analysis of Their Binding Affinity for Omega-Aminocarboxylic Acids Authors: Marti, D. / Schaller, J. / Ochensberger, B. / Rickli, E.E. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1b2i.cif.gz 1b2i.cif.gz | 515.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1b2i.ent.gz pdb1b2i.ent.gz | 430.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1b2i.json.gz 1b2i.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/b2/1b2i https://data.pdbj.org/pub/pdb/validation_reports/b2/1b2i ftp://data.pdbj.org/pub/pdb/validation_reports/b2/1b2i ftp://data.pdbj.org/pub/pdb/validation_reports/b2/1b2i | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 9651.812 Da / Num. of mol.: 1 / Fragment: KRINGLE 2 DOMAIN / Mutation: C181T, E182S, C188G Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Plasmid: PQE-8 / Species (production host): Escherichia coli / Production host: Homo sapiens (human) / Plasmid: PQE-8 / Species (production host): Escherichia coli / Production host:  |
|---|---|
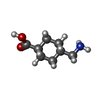
| #2: Chemical | ChemComp-AMH / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 90% H2O/10% D2O |
|---|---|
| Sample conditions | Ionic strength: SEE PAPER / pH: 5.1 / Pressure: AMBIENT / Temperature: 310 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: DISTANCE GEOMETRY, DYNAMIC SIMULATED ANNEALING / Software ordinal: 1 | ||||||||||||
| NMR ensemble | Conformer selection criteria: TOTAL ENERGY, RAMACHANDRAN PLOT, LEAST RESTRAINT VIOLATION Conformers calculated total number: 150 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller









 PDBj
PDBj





 HSQC
HSQC