[English] 日本語
 Yorodumi
Yorodumi- PDB-1akp: SEQUENTIAL 1H,13C AND 15N NMR ASSIGNMENTS AND SOLUTION CONFORMATI... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1akp | ||||||
|---|---|---|---|---|---|---|---|
| Title | SEQUENTIAL 1H,13C AND 15N NMR ASSIGNMENTS AND SOLUTION CONFORMATION OF APOKEDARCIDIN | ||||||
 Components Components | APOKEDARCIDIN | ||||||
 Keywords Keywords | ANTIBIOTIC CHROMOPROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  actinomycete ATCC 53650 (bacteria) actinomycete ATCC 53650 (bacteria) | ||||||
| Method | SOLUTION NMR | ||||||
 Authors Authors | Constantine, K.L. / Colson, K.L. / Wittekind, M. / Friedrichs, M.S. / Zein, N. / Tuttle, J. / Langley, D.R. / Leet, J.E. / Schroeder, D.R. / Lam, K.S. ...Constantine, K.L. / Colson, K.L. / Wittekind, M. / Friedrichs, M.S. / Zein, N. / Tuttle, J. / Langley, D.R. / Leet, J.E. / Schroeder, D.R. / Lam, K.S. / Farmer II, B.T. / Metzler, W.J. / Bruccoleri, R.E. / Mueller, L. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1994 Journal: Biochemistry / Year: 1994Title: Sequential 1H, 13C, and 15N NMR assignments and solution conformation of apokedarcidin. Authors: Constantine, K.L. / Colson, K.L. / Wittekind, M. / Friedrichs, M.S. / Zein, N. / Tuttle, J. / Langley, D.R. / Leet, J.E. / Schroeder, D.R. / Lam, K.S. / Farmer III, B.T. / Metzler, W.J. / ...Authors: Constantine, K.L. / Colson, K.L. / Wittekind, M. / Friedrichs, M.S. / Zein, N. / Tuttle, J. / Langley, D.R. / Leet, J.E. / Schroeder, D.R. / Lam, K.S. / Farmer III, B.T. / Metzler, W.J. / Bruccoleri, R.E. / Mueller, L. #1:  Journal: J.Am.Chem.Soc. / Year: 1993 Journal: J.Am.Chem.Soc. / Year: 1993Title: Chemistry and Structure Elucidation of the Kedarcidin Chromophore Authors: Leet, J.E. / Schroeder, D.R. / Langley, D.R. / Colson, K.L. / Huang, S. / Klohr, S.E. / Lee, M.S. / Golik, J. / Hofstead, S.J. / Doyle, T.W. / Matson, J.A. #2:  Journal: Proc.Natl.Acad.Sci.USA / Year: 1993 Journal: Proc.Natl.Acad.Sci.USA / Year: 1993Title: Kedarcidin Chromophore: An Enediyne that Cleaves DNA in a Sequence-Specific Manner Authors: Zein, N. / Colson, K.L. / Leet, J.E. / Schroeder, D.R. / Solomon, W. / Doyle, T.W. / Casazza, A.M. #3:  Journal: Proc.Natl.Acad.Sci.USA / Year: 1993 Journal: Proc.Natl.Acad.Sci.USA / Year: 1993Title: Selective Proteolytic Activity of the Antitumor Agent Kedarcidin Authors: Zein, N. / Casazza, A.M. / Doyle, T.W. / Leet, J.E. / Scheoeder, D.R. / Solomon, W. / Nadler, S.G. #4:  Journal: J.Antibiot. / Year: 1992 Journal: J.Antibiot. / Year: 1992Title: Kedarcidin, a New Chromoprotein Antitumor Antibiotic. II. Isolation, Purification and Physico-Chemical Properties Authors: Hofstead, S.J. / Matson, J.A. / Malacko, A.R. / Marquart, H. #5:  Journal: J.Antibiot. / Year: 1991 Journal: J.Antibiot. / Year: 1991Title: Kedarcidin, a New Chromoprotein Antitumor Antibiotic I. Taxonomy of Producing Organism, Fermentation and Biological Activity Authors: Lam, K.S. / Hesler, G.A. / Gustavson, D.R. / Crosswell, A.R. / Veitch, J.M. / Forenza, S. / Tomita, K. | ||||||
| History |
|
- Structure visualization
Structure visualization
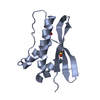
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1akp.cif.gz 1akp.cif.gz | 438 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1akp.ent.gz pdb1akp.ent.gz | 364.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1akp.json.gz 1akp.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1akp_validation.pdf.gz 1akp_validation.pdf.gz | 346.2 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1akp_full_validation.pdf.gz 1akp_full_validation.pdf.gz | 426.4 KB | Display | |
| Data in XML |  1akp_validation.xml.gz 1akp_validation.xml.gz | 22.8 KB | Display | |
| Data in CIF |  1akp_validation.cif.gz 1akp_validation.cif.gz | 43.5 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ak/1akp https://data.pdbj.org/pub/pdb/validation_reports/ak/1akp ftp://data.pdbj.org/pub/pdb/validation_reports/ak/1akp ftp://data.pdbj.org/pub/pdb/validation_reports/ak/1akp | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Atom site foot note | 1: CIS PROLINE - PRO 9 / 2: CIS PROLINE - PRO 81 | |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 10972.936 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  actinomycete ATCC 53650 (bacteria) / Strain: L585-6 / ATCC 53650 / References: UniProt: P41249 actinomycete ATCC 53650 (bacteria) / Strain: L585-6 / ATCC 53650 / References: UniProt: P41249 |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|
- Sample preparation
Sample preparation
| Crystal grow | *PLUS Method: other / Details: NMR |
|---|
- Processing
Processing
| Software |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| NMR software | Name:  X-PLOR / Developer: BRUNGER / Classification: refinement X-PLOR / Developer: BRUNGER / Classification: refinement | ||||||||
| NMR ensemble | Conformers submitted total number: 15 |
 Movie
Movie Controller
Controller











 PDBj
PDBj