[English] 日本語
 Yorodumi
Yorodumi- PDB-1aef: SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACT... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1aef | ||||||
|---|---|---|---|---|---|---|---|
| Title | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (3-AMINOPYRIDINE) | ||||||
 Components Components | CYTOCHROME C PEROXIDASE | ||||||
 Keywords Keywords | OXIDOREDUCTASE / PEROXIDASE / TRANSIT PEPTIDE | ||||||
| Function / homology |  Function and homology information Function and homology informationcytochrome-c peroxidase / cytochrome-c peroxidase activity / response to reactive oxygen species / hydrogen peroxide catabolic process / peroxidase activity / mitochondrial intermembrane space / cellular response to oxidative stress / mitochondrial matrix / heme binding / mitochondrion / metal ion binding Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.1 Å MOLECULAR REPLACEMENT / Resolution: 2.1 Å | ||||||
 Authors Authors | Musah, R.A. / Jensen, G.M. / Fitzgerald, M.M. / Mcree, D.E. / Goodin, D.B. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2002 Journal: J.Mol.Biol. / Year: 2002Title: Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein. Authors: Musah, R.A. / Jensen, G.M. / Bunte, S.W. / Rosenfeld, R.J. / Goodin, D.B. #1:  Journal: Biochemistry / Year: 1994 Journal: Biochemistry / Year: 1994Title: Small Molecule Binding to an Artificially Created Cavity at the Active Site of Cytochrome C Peroxidase Authors: Fitzgerald, M.M. / Churchill, M.J. / Mcree, D.E. / Goodin, D.B. #2:  Journal: Biochemistry / Year: 1993 Journal: Biochemistry / Year: 1993Title: The Asp-His-Fe Triad of Cytochrome C Peroxidase Controls the Reduction Potential, Electronic Structure, and Coupling of the Tryptophan Free Radical to the Heme Authors: Goodin, D.B. / Mcree, D.E. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1aef.cif.gz 1aef.cif.gz | 86 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1aef.ent.gz pdb1aef.ent.gz | 64.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1aef.json.gz 1aef.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ae/1aef https://data.pdbj.org/pub/pdb/validation_reports/ae/1aef ftp://data.pdbj.org/pub/pdb/validation_reports/ae/1aef ftp://data.pdbj.org/pub/pdb/validation_reports/ae/1aef | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1aebC  1aedC  1aeeC  1aegC  1aehC  1aejC  1aekC  1aemC  1aenC  1aeoC  1aeqC  1aa4S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 33459.242 Da / Num. of mol.: 1 / Mutation: W191G Source method: isolated from a genetically manipulated source Details: CRYSTAL FORM BY Source: (gene. exp.)  Cell line: BL21 / Cellular location: MITOCHONDRIA / Gene: CCP / Organelle: MITOCHONDRIA / Plasmid: PT7CCP / Species (production host): Escherichia coli / Cellular location (production host): CYTOPLASM / Gene (production host): CCP (MKT) / Production host:  |
|---|---|
| #2: Chemical | ChemComp-HEM / |
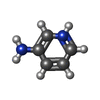
| #3: Chemical | ChemComp-3AP / |
| #4: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.2 Å3/Da / Density % sol: 61 % | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 6 Details: 20% MPD, 40 MM PHOSPHATE PH 6.0. A SINGLE CRYSTAL OF W191G WAS SOAKED IN 50MM 3-AMINOPYRIDINE IN 40% MPD AND 60 MM PHOSPHATE AT PH 4.5. | ||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 18 ℃ / Method: vapor diffusion, sitting drop | ||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 290 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 |
| Detector | Type: SIEMENS / Detector: AREA DETECTOR / Date: Jun 1, 1996 |
| Radiation | Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.1→15 Å / Num. obs: 23265 / % possible obs: 79 % / Observed criterion σ(I): 0 / Redundancy: 1.8 % / Rmerge(I) obs: 0.089 / Rsym value: 0.087 / Net I/σ(I): 11.94 |
| Reflection shell | Resolution: 2.03→2.18 Å / Redundancy: 1.68 % / Rmerge(I) obs: 0.26 / Mean I/σ(I) obs: 1.51 / Rsym value: 0.54 / % possible all: 79 |
| Reflection | *PLUS Highest resolution: 2 Å / Lowest resolution: 15 Å / Rmerge(I) obs: 0.087 |
- Processing
Processing
| Software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1AA4 Resolution: 2.1→7 Å / Data cutoff high absF: 10000000 / Data cutoff low absF: 0.5 / σ(F): 0 Details: THE PROTEIN AND WATER COORDINATES WERE TAKEN FROM PDB ENTRY 1AA4. THE LIGAND COORDINATES WERE MEASURED FROM THE OMIT MAP. NO REFINEMENT WAS CARRIED OUT ON THE RESULTING SET OF COORDINATES, ...Details: THE PROTEIN AND WATER COORDINATES WERE TAKEN FROM PDB ENTRY 1AA4. THE LIGAND COORDINATES WERE MEASURED FROM THE OMIT MAP. NO REFINEMENT WAS CARRIED OUT ON THE RESULTING SET OF COORDINATES, WHICH ARE THE COORDINATES PRESENTED IN THIS ENTRY.
| ||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.1→7 Å
|
 Movie
Movie Controller
Controller






















 PDBj
PDBj