[English] 日本語
 Yorodumi
Yorodumi- PDB-106d: Solution structures of the i-motif tetramers of D(TCC), D(5MCCT) ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 106d | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Solution structures of the i-motif tetramers of D(TCC), D(5MCCT) and D(T5MCC). Novel NOE connections between amino protons and sugar protons | ||||||||||||||||||
 Components Components | DNA (5'-D(* Keywords KeywordsDNA | Function / homology | DNA |  Function and homology information Function and homology informationMethod | SOLUTION NMR |  Authors AuthorsLeroy, J.-L. / Gueron, M. |  Citation Citation Journal: Structure / Year: 1995 Journal: Structure / Year: 1995Title: Solution structures of the i-motif tetramers of d(TCC), d(5methylCCT) and d(T5methylCC): novel NOE connections between amino protons and sugar protons. Authors: Leroy, J.L. / Gueron, M. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  106d.cif.gz 106d.cif.gz | 65.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb106d.ent.gz pdb106d.ent.gz | 40.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  106d.json.gz 106d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  106d_validation.pdf.gz 106d_validation.pdf.gz | 320.2 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  106d_full_validation.pdf.gz 106d_full_validation.pdf.gz | 405.6 KB | Display | |
| Data in XML |  106d_validation.xml.gz 106d_validation.xml.gz | 5.9 KB | Display | |
| Data in CIF |  106d_validation.cif.gz 106d_validation.cif.gz | 7.3 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/06/106d https://data.pdbj.org/pub/pdb/validation_reports/06/106d ftp://data.pdbj.org/pub/pdb/validation_reports/06/106d ftp://data.pdbj.org/pub/pdb/validation_reports/06/106d | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 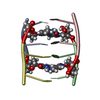
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Atom site foot note | 1: SOME OF THE CYTIDINES ARE PROTONATED AT THE N3 POSITION. THESE RESIDUES HAVE BEEN ASSIGNED THE RESIDUE NAME C IN THE ENTRY AND ATOM H3 HAS BEEN FOOTNOTED. | |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 771.646 Da / Num. of mol.: 4 / Source method: obtained synthetically |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|
- Sample preparation
Sample preparation
| Crystal grow | *PLUS Method: other / Details: NMR |
|---|
- Processing
Processing
| Software |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Refinement | Software ordinal: 1 Details: 5MC STANDS FOR 5-METHYLCYTIDINE. THE STRUCTURE IS FORMED OF FOUR EQUIVALENT 5MCCT STRANDS DESIGNATED A, B, C, D. STRANDS A AND C, CONNECTED BY TWO HEMI-PROTONATED BASE PAIRS (5MC1.5MC1+ AND ...Details: 5MC STANDS FOR 5-METHYLCYTIDINE. THE STRUCTURE IS FORMED OF FOUR EQUIVALENT 5MCCT STRANDS DESIGNATED A, B, C, D. STRANDS A AND C, CONNECTED BY TWO HEMI-PROTONATED BASE PAIRS (5MC1.5MC1+ AND C2.C2+), FORM A PARALLEL-STRANDED DUPLEX. THE RELATION BETWEEN STRANDS B AND D IS THE SAME AS THAT BETWEEN STRANDS A AND C. THE STRANDS OF EACH DUPLEX ARE RELATED BY A LONGITUDINAL TWO-FOLD SYMMETRY AXIS. THE DUPLEXES HAVE THE SAME LONGITUDINAL AXIS, AND THEY ARE ANTI-PARALLEL. THEIR HEMI-PROTONATED C.C+ BASE PAIRS ARE INTERCALATED FACE-TO-FACE. THE DUPLEXES CAN BE TRANSFORMED INTO ONE ANOTHER BY A 180 DEGREE ROTATION AROUND A TWO-FOLD AXIS PERPENDICULAR TO THE LONGITUDINAL AXIS, AND CONSEQUENTLY AROUND A THIRD AXIS PERPENDICULAR TO THE FIRST TWO. THE H3 IMINO PROTONS OF THE HEMI-PROTONATED 5MC1.5MC1+ PAIRS WERE NOT INCORPORATED IN THE COMPUTATIONS. THOSE OF THE C2.C2+ PAIRS WERE ARBITRARILY LOCATED ON THE C2 RESIDUES OF STRANDS A AND B, RESPECTIVELY. | ||||||||
| NMR ensemble | Conformers submitted total number: 8 |
 Movie
Movie Controller
Controller






 PDBj
PDBj

 X-PLOR
X-PLOR