+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9998 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human MLL1-ubNCP complex (3.2 angstrom) | |||||||||
 Map data Map data | Main map of MLL1-ubNCP (3.2 angstrom) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | histone modification / nucleosome / MLL / TRANSCRIPTION / TRANSCRIPTION-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of DNA methylation-dependent heterochromatin formation / [histone H3]-lysine4 N-methyltransferase / histone H3K4 monomethyltransferase activity / protein-cysteine methyltransferase activity / response to potassium ion / histone H3K4 trimethyltransferase activity / unmethylated CpG binding / histone H3Q5ser reader activity / histone H3K4me1 reader activity / Loss of Function of KMT2D in MLL4 Complex Formation in Kabuki Syndrome ...negative regulation of DNA methylation-dependent heterochromatin formation / [histone H3]-lysine4 N-methyltransferase / histone H3K4 monomethyltransferase activity / protein-cysteine methyltransferase activity / response to potassium ion / histone H3K4 trimethyltransferase activity / unmethylated CpG binding / histone H3Q5ser reader activity / histone H3K4me1 reader activity / Loss of Function of KMT2D in MLL4 Complex Formation in Kabuki Syndrome / Epigenetic regulation of gene expression by MLL3 and MLL4 complexes / MLL3/4 complex / Set1C/COMPASS complex / definitive hemopoiesis / ATAC complex / MLL1/2 complex / NSL complex / regulation of short-term neuronal synaptic plasticity / histone H3K4 methyltransferase activity / Cardiogenesis / anterior/posterior pattern specification / embryonic hemopoiesis / T-helper 2 cell differentiation / Formation of the ternary complex, and subsequently, the 43S complex / Formation of WDR5-containing histone-modifying complexes / Ribosomal scanning and start codon recognition / exploration behavior / Translation initiation complex formation / histone methyltransferase complex / minor groove of adenine-thymine-rich DNA binding / hemopoiesis / MLL1 complex / SARS-CoV-1 modulates host translation machinery / regulation of embryonic development / histone acetyltransferase complex / regulation of cell division / Peptide chain elongation / Selenocysteine synthesis / Formation of a pool of free 40S subunits / membrane depolarization / Eukaryotic Translation Termination / SRP-dependent cotranslational protein targeting to membrane / Response of EIF2AK4 (GCN2) to amino acid deficiency / Viral mRNA Translation / cellular response to transforming growth factor beta stimulus / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / GTP hydrolysis and joining of the 60S ribosomal subunit / L13a-mediated translational silencing of Ceruloplasmin expression / Major pathway of rRNA processing in the nucleolus and cytosol / negative regulation of fibroblast proliferation / spleen development / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / Maturation of protein E / Maturation of protein E / ER Quality Control Compartment (ERQC) / Myoclonic epilepsy of Lafora / FLT3 signaling by CBL mutants / Constitutive Signaling by NOTCH1 HD Domain Mutants / IRAK2 mediated activation of TAK1 complex / Prevention of phagosomal-lysosomal fusion / Alpha-protein kinase 1 signaling pathway / Glycogen synthesis / IRAK1 recruits IKK complex / IRAK1 recruits IKK complex upon TLR7/8 or 9 stimulation / Endosomal Sorting Complex Required For Transport (ESCRT) / homeostasis of number of cells within a tissue / Membrane binding and targetting of GAG proteins / positive regulation of gluconeogenesis / Negative regulation of FLT3 / Regulation of TBK1, IKKε (IKBKE)-mediated activation of IRF3, IRF7 / PTK6 Regulates RTKs and Their Effectors AKT1 and DOK1 / transcription initiation-coupled chromatin remodeling / Regulation of TBK1, IKKε-mediated activation of IRF3, IRF7 upon TLR3 ligation / IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation / NOTCH2 Activation and Transmission of Signal to the Nucleus / TICAM1,TRAF6-dependent induction of TAK1 complex / TICAM1-dependent activation of IRF3/IRF7 / APC/C:Cdc20 mediated degradation of Cyclin B / Regulation of FZD by ubiquitination / Downregulation of ERBB4 signaling / APC-Cdc20 mediated degradation of Nek2A / p75NTR recruits signalling complexes / cytosolic ribosome / InlA-mediated entry of Listeria monocytogenes into host cells / TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling / Regulation of pyruvate metabolism / NF-kB is activated and signals survival / TRAF6-mediated induction of TAK1 complex within TLR4 complex / Downregulation of ERBB2:ERBB3 signaling / Pexophagy / Regulation of innate immune responses to cytosolic DNA / sperm principal piece / NRIF signals cell death from the nucleus / Regulation of PTEN localization / Activated NOTCH1 Transmits Signal to the Nucleus / Transferases; Transferring one-carbon groups; Methyltransferases / VLDLR internalisation and degradation / Synthesis of active ubiquitin: roles of E1 and E2 enzymes / TICAM1, RIP1-mediated IKK complex recruitment / Regulation of BACH1 activity Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) / | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Huang J / Xue H | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: Structural basis of nucleosome recognition and modification by MLL methyltransferases. Authors: Han Xue / Tonghui Yao / Mi Cao / Guanjun Zhu / Yan Li / Guiyong Yuan / Yong Chen / Ming Lei / Jing Huang /  Abstract: Methyltransferases of the mixed-lineage leukaemia (MLL) family-which include MLL1, MLL2, MLL3, MLL4, SET1A and SET1B-implement methylation of histone H3 on lysine 4 (H3K4), and have critical and ...Methyltransferases of the mixed-lineage leukaemia (MLL) family-which include MLL1, MLL2, MLL3, MLL4, SET1A and SET1B-implement methylation of histone H3 on lysine 4 (H3K4), and have critical and distinct roles in the regulation of transcription in haematopoiesis, adipogenesis and development. The C-terminal catalytic SET (Su(var.)3-9, enhancer of zeste and trithorax) domains of MLL proteins are associated with a common set of regulatory factors (WDR5, RBBP5, ASH2L and DPY30) to achieve specific activities. Current knowledge of the regulation of MLL activity is limited to the catalysis of histone H3 peptides, and how H3K4 methyl marks are deposited on nucleosomes is poorly understood. H3K4 methylation is stimulated by mono-ubiquitination of histone H2B on lysine 120 (H2BK120ub1), a prevalent histone H2B mark that disrupts chromatin compaction and favours open chromatin structures, but the underlying mechanism remains unknown. Here we report cryo-electron microscopy structures of human MLL1 and MLL3 catalytic modules associated with nucleosome core particles that contain H2BK120ub1 or unmodified H2BK120. These structures demonstrate that the MLL1 and MLL3 complexes both make extensive contacts with the histone-fold and DNA regions of the nucleosome; this allows ease of access to the histone H3 tail, which is essential for the efficient methylation of H3K4. The H2B-conjugated ubiquitin binds directly to RBBP5, orienting the association between MLL1 or MLL3 and the nucleosome. The MLL1 and MLL3 complexes display different structural organizations at the interface between the WDR5, RBBP5 and MLL1 (or the corresponding MLL3) subunits, which accounts for the opposite roles of WDR5 in regulating the activity of the two enzymes. These findings transform our understanding of the structural basis for the regulation of MLL activity at the nucleosome level, and highlight the pivotal role of nucleosome regulation in histone-tail modification. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9998.map.gz emd_9998.map.gz | 69.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9998-v30.xml emd-9998-v30.xml emd-9998.xml emd-9998.xml | 34 KB 34 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9998_fsc.xml emd_9998_fsc.xml | 9.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_9998.png emd_9998.png | 209.2 KB | ||
| Masks |  emd_9998_msk_1.map emd_9998_msk_1.map | 75.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-9998.cif.gz emd-9998.cif.gz | 7.9 KB | ||
| Others |  emd_9998_additional_1.map.gz emd_9998_additional_1.map.gz emd_9998_additional_2.map.gz emd_9998_additional_2.map.gz emd_9998_additional_3.map.gz emd_9998_additional_3.map.gz emd_9998_additional_4.map.gz emd_9998_additional_4.map.gz | 70 MB 70 MB 69.9 MB 69.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9998 http://ftp.pdbj.org/pub/emdb/structures/EMD-9998 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9998 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9998 | HTTPS FTP |
-Related structure data
| Related structure data |  6kiuMC  0693C  0694C  0695C  9999C  6kivC  6kiwC  6kixC  6kizC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9998.map.gz / Format: CCP4 / Size: 75.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9998.map.gz / Format: CCP4 / Size: 75.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map of MLL1-ubNCP (3.2 angstrom) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
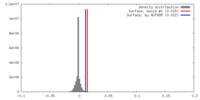
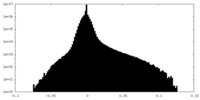
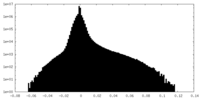
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_9998_msk_1.map emd_9998_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
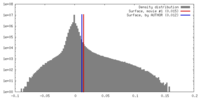
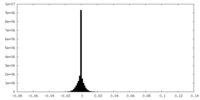
| Density Histograms |
-Additional map: Additional map showing good density of RBBP5 AS
| File | emd_9998_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional map showing good density of RBBP5_AS | ||||||||||||
| Projections & Slices |
| ||||||||||||
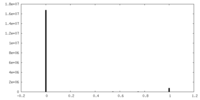
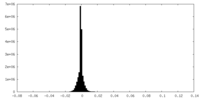
| Density Histograms |
-Additional map: Additional map showing good density of MLL1 AS
| File | emd_9998_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional map showing good density of MLL1_AS | ||||||||||||
| Projections & Slices |
| ||||||||||||
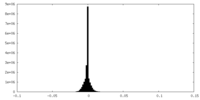
| Density Histograms |
-Additional map: Additional map showing good density of H2BK120ub1
| File | emd_9998_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional map showing good density of H2BK120ub1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Additional map showing good density of ASH2L SPRY
| File | emd_9998_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional map showing good density of ASH2L_SPRY | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Human MLL1 complex associated with an H2B-monoubiquitinated nucle...
+Supramolecule #1: Human MLL1 complex associated with an H2B-monoubiquitinated nucle...
+Supramolecule #2: Human MLL1
+Supramolecule #3: H2B-monoubiquitinated nucleosome
+Macromolecule #1: Histone H3
+Macromolecule #2: Histone H4
+Macromolecule #3: Histone H2A
+Macromolecule #4: Histone H2B 1.1
+Macromolecule #7: Histone-lysine N-methyltransferase 2A
+Macromolecule #8: Retinoblastoma-binding protein 5
+Macromolecule #9: WD repeat-containing protein 5
+Macromolecule #10: Set1/Ash2 histone methyltransferase complex subunit ASH2
+Macromolecule #11: Ubiquitin
+Macromolecule #5: DNA (145-MER)
+Macromolecule #6: DNA (145-MER)
+Macromolecule #12: S-ADENOSYL-L-HOMOCYSTEINE
+Macromolecule #13: ZINC ION
+Macromolecule #14: LYSINE
+Macromolecule #15: GLUTAMINE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)