+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9953 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural insights into stressosome assembly | |||||||||
 Map data Map data | RsbRA/S complex | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
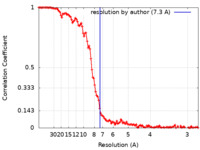
| Method | single particle reconstruction / cryo EM / Resolution: 7.3 Å | |||||||||
 Authors Authors | Kim H / Kwon E / Pathak D / Kim DY / Jung HS | |||||||||
| Funding support |  Korea, Republic Of, 2 items Korea, Republic Of, 2 items
| |||||||||
 Citation Citation | Journal: J Mol Biol / Year: 2011 Title: Role of the tail in the regulated state of myosin 2. Authors: Hyun Suk Jung / Neil Billington / Kavitha Thirumurugan / Bridget Salzameda / Christine R Cremo / Joseph M Chalovich / Peter D Chantler / Peter J Knight /  Abstract: Myosin 2 from vertebrate smooth muscle or non-muscle sources is in equilibrium between compact, inactive monomers and thick filaments under physiological conditions. In the inactive monomer, the two ...Myosin 2 from vertebrate smooth muscle or non-muscle sources is in equilibrium between compact, inactive monomers and thick filaments under physiological conditions. In the inactive monomer, the two heads pack compactly together, and the long tail is folded into three closely packed segments that are associated chiefly with one of the heads. The molecular basis of the folding of the tail remains unexplained. By using electron microscopy, we show that compact monomers of smooth muscle myosin 2 have the same structure in both the native state and following specific, intramolecular photo-cross-linking between Cys109 of the regulatory light chain (RLC) and segment 3 of the tail. Nonspecific cross-linking between lysine residues of the folded monomer by glutaraldehyde also does not perturb the compact conformation and stabilizes it against unfolding at high ionic strength. Sequence comparisons across phyla and myosin 2 isoforms suggest that the folding of the tail is stabilized by ionic interactions between the positively charged N-terminal sequence of the RLC and a negatively charged region near the start of tail segment 3 and that phosphorylation of the RLC could perturb these interactions. Our results support the view that interactions between the heads and the distal tail perform a critical role in regulating activity of myosin 2 molecules through stabilizing the compact monomer conformation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9953.map.gz emd_9953.map.gz | 10.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9953-v30.xml emd-9953-v30.xml emd-9953.xml emd-9953.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9953_fsc.xml emd_9953_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_9953.png emd_9953.png | 206.8 KB | ||
| Masks |  emd_9953_msk_1.map emd_9953_msk_1.map | 137.1 MB |  Mask map Mask map | |
| Others |  emd_9953_half_map_1.map.gz emd_9953_half_map_1.map.gz emd_9953_half_map_2.map.gz emd_9953_half_map_2.map.gz | 105.8 MB 105.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9953 http://ftp.pdbj.org/pub/emdb/structures/EMD-9953 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9953 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9953 | HTTPS FTP |
-Validation report
| Summary document |  emd_9953_validation.pdf.gz emd_9953_validation.pdf.gz | 447.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9953_full_validation.pdf.gz emd_9953_full_validation.pdf.gz | 447.4 KB | Display | |
| Data in XML |  emd_9953_validation.xml.gz emd_9953_validation.xml.gz | 19.1 KB | Display | |
| Data in CIF |  emd_9953_validation.cif.gz emd_9953_validation.cif.gz | 25 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9953 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9953 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9953 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9953 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9953.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9953.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RsbRA/S complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
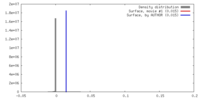
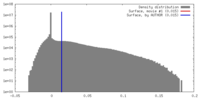
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
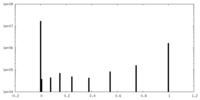
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_9953_msk_1.map emd_9953_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_9953_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_9953_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : RsbRA/RsbS complex from Bacillus subtilis
| Entire | Name: RsbRA/RsbS complex from Bacillus subtilis |
|---|---|
| Components |
|
-Supramolecule #1: RsbRA/RsbS complex from Bacillus subtilis
| Supramolecule | Name: RsbRA/RsbS complex from Bacillus subtilis / type: complex / ID: 1 / Parent: 0 Details: Stressosome (RsbRA/RsbS) complex from Bacillus subtilis |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Enterobacteria phage L1 (virus) / Recombinant strain: BL-21star (DE3) Enterobacteria phage L1 (virus) / Recombinant strain: BL-21star (DE3) |
| Molecular weight | Experimental: 1.8 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 20 mM HEPES, 50 mM NaCl, 0.2 mM TCEP, 5 % glycerol, pH 7.5 | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 39.2 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 7 seconds before plunging. | |||||||||||||||
| Details | RsbRA/RsbS complex buffer: 20 mM HEPES, 50 mM NaCl, 0.2 mM TCEP, 5 % glycerol, pH 7.5 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: OTHER / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number real images: 1788 / Average exposure time: 1.0 sec. / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.0 µm / Calibrated defocus min: 1.5 µm / Calibrated magnification: 47000 / Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 47000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)