+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9730 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Flock House Virus eluted particle | |||||||||
 Map data Map data | Cryo-EM structure of Flock House Virus eluted particle | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Flock House Virus / Disassembly intermediate / Asymmetric reconstruction / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationnodavirus endopeptidase / symbiont entry into host cell via permeabilization of host membrane / T=3 icosahedral viral capsid / aspartic-type endopeptidase activity / proteolysis / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Flock house virus Flock house virus | |||||||||
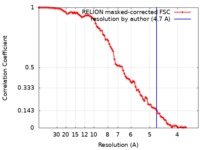
| Method | single particle reconstruction / cryo EM / Resolution: 4.7 Å | |||||||||
 Authors Authors | Banerjee M / Azad K | |||||||||
| Funding support |  India, 1 items India, 1 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2019 Journal: J Virol / Year: 2019Title: Structural Dynamics of Nonenveloped Virus Disassembly Intermediates. Authors: Kimi Azad / Manidipa Banerjee /  Abstract: The stability of icosahedral viruses is crucial for protecting the viral genome during transit; however, successful infection requires eventual disassembly of the capsid. A comprehensive ...The stability of icosahedral viruses is crucial for protecting the viral genome during transit; however, successful infection requires eventual disassembly of the capsid. A comprehensive understanding of how stable, uniform icosahedrons disassemble remains elusive, mainly due to the complexities involved in isolating transient intermediates. We utilized incremental heating to systematically characterize the disassembly pathway of a model nonenveloped virus and identified an intriguing link between virus maturation and disassembly. Further, we isolated and characterized two intermediates by cryo-electron microscopy and three-dimensional reconstruction, without imposing icosahedral symmetry. The first intermediate displayed a series of major, asymmetric alterations, whereas the second showed that the act of genome release, through the 2-fold axis, is actually confined to a small section on the capsid. Our study thus presents a comprehensive structural analysis of nonenveloped virus disassembly and emphasizes the asymmetric nature of programmed conformational changes. Disassembly or uncoating of an icosahedral capsid is a crucial step during infection by nonenveloped viruses. However, the dynamic and transient nature of the disassembly process makes it challenging to isolate intermediates in a temporal, stepwise manner for structural characterization. Using controlled, incremental heating, we isolated two disassembly intermediates: "eluted particles" and "puffed particles" of an insect nodavirus, Flock House virus (FHV). Cryo-electron microscopy and three-dimensional reconstruction of the FHV disassembly intermediates indicated that disassembly-related conformational alterations are minimally global and largely local, leading to asymmetry in the particle and eventual genome release without complete disintegration of the icosahedron. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9730.map.gz emd_9730.map.gz | 15.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9730-v30.xml emd-9730-v30.xml emd-9730.xml emd-9730.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9730_fsc.xml emd_9730_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_9730.png emd_9730.png | 197.2 KB | ||
| Filedesc metadata |  emd-9730.cif.gz emd-9730.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9730 http://ftp.pdbj.org/pub/emdb/structures/EMD-9730 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9730 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9730 | HTTPS FTP |
-Validation report
| Summary document |  emd_9730_validation.pdf.gz emd_9730_validation.pdf.gz | 594.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9730_full_validation.pdf.gz emd_9730_full_validation.pdf.gz | 594.4 KB | Display | |
| Data in XML |  emd_9730_validation.xml.gz emd_9730_validation.xml.gz | 11.1 KB | Display | |
| Data in CIF |  emd_9730_validation.cif.gz emd_9730_validation.cif.gz | 14.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9730 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9730 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9730 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9730 | HTTPS FTP |
-Related structure data
| Related structure data |  6itbMC  6itfMC  9732C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9730.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9730.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of Flock House Virus eluted particle | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.7407 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
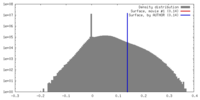
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Flock house virus
| Entire | Name:  Flock house virus Flock house virus |
|---|---|
| Components |
|
-Supramolecule #1: Flock house virus
| Supramolecule | Name: Flock house virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Purified wildtype Flock House Virus heated to 70 degC NCBI-ID: 12287 / Sci species name: Flock house virus / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Costelytra zealandica (beetle) Costelytra zealandica (beetle) |
| Molecular weight | Theoretical: 9 MDa |
| Virus shell | Shell ID: 1 / Diameter: 300.0 Å / T number (triangulation number): 3 |
-Macromolecule #1: CAPSID PROTEIN BETA
| Macromolecule | Name: CAPSID PROTEIN BETA / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO / EC number: nodavirus endopeptidase |
|---|---|
| Source (natural) | Organism:  Flock house virus Flock house virus |
| Molecular weight | Theoretical: 39.367355 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVNNNRPRRQ RAQRVVVTTT QTAPVPQQNV PRNGRRRRNR TRRNRRRVRG MNMAALTRLS QPGLAFLKCA FAPPDFNTDP GKGIPDRFE GKVVSRKDVL NQSISFTAGQ DTFILIAPTP GVAYWSASVP AGTFPTSATT FNPVNYPGFT SMFGTTSTSR S DQVSSFRY ...String: MVNNNRPRRQ RAQRVVVTTT QTAPVPQQNV PRNGRRRRNR TRRNRRRVRG MNMAALTRLS QPGLAFLKCA FAPPDFNTDP GKGIPDRFE GKVVSRKDVL NQSISFTAGQ DTFILIAPTP GVAYWSASVP AGTFPTSATT FNPVNYPGFT SMFGTTSTSR S DQVSSFRY ASMNVGIYPT SNLMQFAGSI TVWKCPVKLS TVQFPVATDP ATSSLVHTLV GLDGVLAVGP DNFSESFIKG VF SQSACNE PDFEFNDILE GIQTLPPANV SLGSTGQPFT MDSGAEATSG VVGWGNMDTI VIRVSAPEGA VNSAILKAWS CIE YRPNPN AMLYQFGHDS PPLDEVALQE YRTVARSLPV AVIAAQN UniProtKB: Capsid protein alpha |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7 / Component - Concentration: 50.0 mM / Component - Formula: C8H18N2O4S / Component - Name: HEPES |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 284 K / Instrument: FEI VITROBOT MARK IV / Details: Blot for 3 seconds before plunging. |
| Details | Purified wildtype Flock House Virus heated to 70 degC |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 2 / Number real images: 2835 / Average exposure time: 5.0 sec. / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 181 |
| Output model |  PDB-6itb:  PDB-6itf: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)