[English] 日本語
 Yorodumi
Yorodumi- EMDB-9350: SegA-asym, conformation of TDP-43 low complexity domain segment A asym -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9350 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SegA-asym, conformation of TDP-43 low complexity domain segment A asym | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Amyloid / TDP43 / ALS / FTLD-TDP / dna binding protein / protein fibril | |||||||||
| Function / homology |  Function and homology information Function and homology informationnuclear inner membrane organization / interchromatin granule / perichromatin fibrils / 3'-UTR-mediated mRNA destabilization / 3'-UTR-mediated mRNA stabilization / intracellular membraneless organelle / negative regulation of protein phosphorylation / host-mediated suppression of viral transcription / pre-mRNA intronic binding / RNA splicing ...nuclear inner membrane organization / interchromatin granule / perichromatin fibrils / 3'-UTR-mediated mRNA destabilization / 3'-UTR-mediated mRNA stabilization / intracellular membraneless organelle / negative regulation of protein phosphorylation / host-mediated suppression of viral transcription / pre-mRNA intronic binding / RNA splicing / response to endoplasmic reticulum stress / mRNA 3'-UTR binding / molecular condensate scaffold activity / regulation of circadian rhythm / positive regulation of insulin secretion / regulation of protein stability / positive regulation of protein import into nucleus / mRNA processing / cytoplasmic stress granule / rhythmic process / regulation of gene expression / double-stranded DNA binding / regulation of apoptotic process / amyloid fibril formation / regulation of cell cycle / nuclear speck / RNA polymerase II cis-regulatory region sequence-specific DNA binding / negative regulation of gene expression / lipid binding / chromatin / mitochondrion / DNA binding / RNA binding / nucleoplasm / identical protein binding / nucleus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Cao Q / Boyer DR | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2019 Journal: Nat Struct Mol Biol / Year: 2019Title: Cryo-EM structures of four polymorphic TDP-43 amyloid cores. Authors: Qin Cao / David R Boyer / Michael R Sawaya / Peng Ge / David S Eisenberg /  Abstract: The DNA and RNA processing protein TDP-43 undergoes both functional and pathogenic aggregation. Functional TDP-43 aggregates form reversible, transient species such as nuclear bodies, stress ...The DNA and RNA processing protein TDP-43 undergoes both functional and pathogenic aggregation. Functional TDP-43 aggregates form reversible, transient species such as nuclear bodies, stress granules, and myo-granules. Pathogenic, irreversible TDP-43 aggregates form in amyotrophic lateral sclerosis and other neurodegenerative conditions. Here we find the features of TDP-43 fibrils that confer both reversibility and irreversibility by determining structures of two segments reported to be the pathogenic cores of human TDP-43 aggregation: SegA (residues 311-360), which forms three polymorphs, all with dagger-shaped folds; and SegB A315E (residues 286-331 containing the amyotrophic lateral sclerosis hereditary mutation A315E), which forms R-shaped folds. Energetic analysis suggests that the dagger-shaped polymorphs represent irreversible fibril structures, whereas the SegB polymorph may participate in both reversible and irreversible fibrils. Our structures reveal the polymorphic nature of TDP-43 and suggest how the A315E mutation converts the R-shaped polymorph to an irreversible form that enhances pathology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9350.map.gz emd_9350.map.gz | 7.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9350-v30.xml emd-9350-v30.xml emd-9350.xml emd-9350.xml | 10.2 KB 10.2 KB | Display Display |  EMDB header EMDB header |
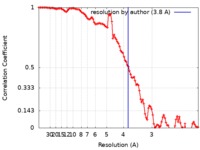
| FSC (resolution estimation) |  emd_9350_fsc.xml emd_9350_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_9350.png emd_9350.png | 165.9 KB | ||
| Filedesc metadata |  emd-9350.cif.gz emd-9350.cif.gz | 5.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9350 http://ftp.pdbj.org/pub/emdb/structures/EMD-9350 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9350 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9350 | HTTPS FTP |
-Validation report
| Summary document |  emd_9350_validation.pdf.gz emd_9350_validation.pdf.gz | 611.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9350_full_validation.pdf.gz emd_9350_full_validation.pdf.gz | 611.4 KB | Display | |
| Data in XML |  emd_9350_validation.xml.gz emd_9350_validation.xml.gz | 11 KB | Display | |
| Data in CIF |  emd_9350_validation.cif.gz emd_9350_validation.cif.gz | 14.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9350 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9350 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9350 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9350 | HTTPS FTP |
-Related structure data
| Related structure data |  6n3bMC  0334C  9339C  9349C  6n37C  6n3aC  6n3cC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10497 (Title: Cryo-EM structures of four polymorphic TDP-43 amyloid cores EMPIAR-10497 (Title: Cryo-EM structures of four polymorphic TDP-43 amyloid coresData size: 4.5 TB Data #1: Unaligned dose fractionated frames of TDP-43 SegA amyloid fibrils [micrographs - multiframe] Data #2: Unaligned dose fractionated frames of TDP-43 SegB A315E amyloid fibrils [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9350.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9350.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.064 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : TDP43 fibril
| Entire | Name: TDP43 fibril |
|---|---|
| Components |
|
-Supramolecule #1: TDP43 fibril
| Supramolecule | Name: TDP43 fibril / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: TAR DNA-binding protein 43
| Macromolecule | Name: TAR DNA-binding protein 43 / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 5.207797 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNFGAFSINP AMMAAAQAAL QSSWGMMGML ASQQNQSGPS GNNQNQGNMQ UniProtKB: TAR DNA-binding protein 43 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average exposure time: 8.0 sec. / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 0.002 µm / Nominal defocus min: 0.002 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6n3b: |
 Movie
Movie Controller
Controller











 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)