[English] 日本語
 Yorodumi
Yorodumi- EMDB-9127: A nucleosome bridging mechanism for activation of a maintenance D... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9127 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | A nucleosome bridging mechanism for activation of a maintenance DNA methyltransferase | |||||||||
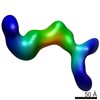
 Map data Map data | Negative stain map of ZMET2 in complex with H3Kc9me3 dinucleosome with 20 pase pairs of linker DNA. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 28.0 Å | |||||||||
 Authors Authors | Stoddard CI / Feng S / Campbell MG / Liu W / Wang H / Zhong X / Bernatavichute Y / Cheng Y / Jacobsen SE / Narlikar GJ | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2019 Journal: Mol Cell / Year: 2019Title: A Nucleosome Bridging Mechanism for Activation of a Maintenance DNA Methyltransferase. Authors: Caitlin I Stoddard / Suhua Feng / Melody G Campbell / Wanlu Liu / Haifeng Wang / Xuehua Zhong / Yana Bernatavichute / Yifan Cheng / Steven E Jacobsen / Geeta J Narlikar /  Abstract: DNA methylation and H3K9me are hallmarks of heterochromatin in plants and mammals, and are successfully maintained across generations. The biochemical and structural basis for this maintenance is ...DNA methylation and H3K9me are hallmarks of heterochromatin in plants and mammals, and are successfully maintained across generations. The biochemical and structural basis for this maintenance is poorly understood. The maintenance DNA methyltransferase from Zea mays, ZMET2, recognizes dimethylation of H3K9 via a chromodomain (CD) and a bromo adjacent homology (BAH) domain, which flank the catalytic domain. Here, we show that dinucleosomes are the preferred ZMET2 substrate, with DNA methylation preferentially targeted to linker DNA. Electron microscopy shows one ZMET2 molecule bridging two nucleosomes within a dinucleosome. We find that the CD stabilizes binding, whereas the BAH domain enables allosteric activation by the H3K9me mark. ZMET2 further couples recognition of H3K9me to an increase in the specificity for hemimethylated versus unmethylated DNA. We propose a model in which synergistic coupling between recognition of nucleosome spacing, H3K9 methylation, and DNA modification allows ZMET2 to maintain DNA methylation in heterochromatin with high fidelity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9127.map.gz emd_9127.map.gz | 1.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9127-v30.xml emd-9127-v30.xml emd-9127.xml emd-9127.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9127.png emd_9127.png | 21 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9127 http://ftp.pdbj.org/pub/emdb/structures/EMD-9127 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9127 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9127 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9127.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9127.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stain map of ZMET2 in complex with H3Kc9me3 dinucleosome with 20 pase pairs of linker DNA. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.14 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of ZMET2 with H3Kc9me3 dinucleosome with 20 base pairs of...
| Entire | Name: Complex of ZMET2 with H3Kc9me3 dinucleosome with 20 base pairs of linker DNA. |
|---|---|
| Components |
|
-Supramolecule #1: Complex of ZMET2 with H3Kc9me3 dinucleosome with 20 base pairs of...
| Supramolecule | Name: Complex of ZMET2 with H3Kc9me3 dinucleosome with 20 base pairs of linker DNA. type: complex / ID: 1 / Parent: 0 |
|---|
-Supramolecule #2: Xenopus histones
| Supramolecule | Name: Xenopus histones / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism: |
| Recombinant expression | Organism:  |
-Supramolecule #3: ZMET2
| Supramolecule | Name: ZMET2 / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Supramolecule #4: 601 dinucleosomal DNA
| Supramolecule | Name: 601 dinucleosomal DNA / type: complex / ID: 4 / Parent: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Formate |
| Grid | Details: unspecified |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)