+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9064 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
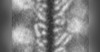
| Title | Cryo-EM structure of the PYD filament of AIM2 | |||||||||
 Map data Map data | PYD filament of AIM2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Filament / higher order / innate immunity / inflammasome / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationpyroptosome complex assembly / AIM2 inflammasome complex assembly / The AIM2 inflammasome / AIM2 inflammasome complex / regulation of behavior / cysteine-type endopeptidase activator activity / Cytosolic sensors of pathogen-associated DNA / pattern recognition receptor signaling pathway / : / pattern recognition receptor activity ...pyroptosome complex assembly / AIM2 inflammasome complex assembly / The AIM2 inflammasome / AIM2 inflammasome complex / regulation of behavior / cysteine-type endopeptidase activator activity / Cytosolic sensors of pathogen-associated DNA / pattern recognition receptor signaling pathway / : / pattern recognition receptor activity / T cell homeostasis / pyroptotic inflammatory response / cellular response to interferon-beta / positive regulation of defense response to virus by host / signaling adaptor activity / activation of innate immune response / negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / bioluminescence / tumor necrosis factor-mediated signaling pathway / positive regulation of interleukin-1 beta production / generation of precursor metabolites and energy / neuron cellular homeostasis / : / brain development / cellular response to xenobiotic stimulus / positive regulation of inflammatory response / site of double-strand break / double-stranded DNA binding / defense response to virus / immune response / innate immune response / DNA damage response / mitochondrion / nucleoplasm / identical protein binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 5.0 Å | |||||||||
 Authors Authors | Lu A / Li Y | |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2015 Journal: Cell Discov / Year: 2015Title: Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2. Authors: Alvin Lu / Yang Li / Qian Yin / Jianbin Ruan / Xiong Yu / Edward Egelman / Hao Wu /  Abstract: Absent in melanoma 2 (AIM2) is an essential cytosolic double-stranded DNA receptor that assembles with the adaptor, apoptosis-associated speck-like protein containing a caspase recruitment domain ...Absent in melanoma 2 (AIM2) is an essential cytosolic double-stranded DNA receptor that assembles with the adaptor, apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC), and caspase-1 to form the AIM2 inflammasome, which leads to proteolytic maturation of cytokines and pyroptotic cell death. AIM2 contains an N-terminal Pyrin domain (PYD) that interacts with ASC through PYD/PYD interactions and nucleates ASC filament formation. To elucidate the molecular basis of AIM2-induced ASC polymerization, we generated AIM2 filaments fused to green fluorescent protein (GFP) and determined its cryo-electron microscopic (cryo-EM) structure. The map showed distinct definition of helices, allowing fitting of the crystal structure. Surprisingly, the GFP-AIM2 filament is a 1-start helix with helical parameters distinct from those of the 3-start ASC filament. However, despite the apparent symmetry difference, helical net and detailed interface analyses reveal minimal changes in subunit packing. GFP-AIM2 nucleated ASC filament formation in comparable efficiency as untagged AIM2, suggesting assembly plasticity in both AIM2 and ASC. The DNA-binding domain of AIM2 is able to form AIM2/DNA filaments, within which the AIM2 is brought into proximity to template ASC filament assembly. Because ASC is able to interact with many PYD-containing receptors for the formation of inflammasomes, the observed structural plasticity may be critically important for this versatility in the PYD/PYD interactions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9064.map.gz emd_9064.map.gz | 20.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9064-v30.xml emd-9064-v30.xml emd-9064.xml emd-9064.xml | 9.9 KB 9.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9064.png emd_9064.png | 70.4 KB | ||
| Filedesc metadata |  emd-9064.cif.gz emd-9064.cif.gz | 5.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9064 http://ftp.pdbj.org/pub/emdb/structures/EMD-9064 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9064 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9064 | HTTPS FTP |
-Related structure data
| Related structure data |  6mb2MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9064.map.gz / Format: CCP4 / Size: 112.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9064.map.gz / Format: CCP4 / Size: 112.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PYD filament of AIM2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.575 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
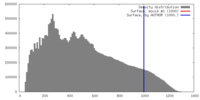
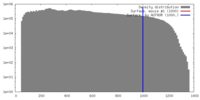
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : PYD of AIM2
| Entire | Name: PYD of AIM2 |
|---|---|
| Components |
|
-Supramolecule #1: PYD of AIM2
| Supramolecule | Name: PYD of AIM2 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Interferon-inducible protein AIM2
| Macromolecule | Name: Interferon-inducible protein AIM2 / type: protein_or_peptide / ID: 1 / Number of copies: 15 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 10.839629 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AMESKYKEIL LLTGLDNITD EELDRFKGFL SDEFNIATGK LHTANRIQVA TLMIQNAGAV SAVMKTIRIF QKLNYMLLAK RLQEEKEKV DKQYK UniProtKB: Interferon-inducible protein AIM2 |
-Macromolecule #2: Green fluorescent protein
| Macromolecule | Name: Green fluorescent protein / type: protein_or_peptide / ID: 2 / Number of copies: 15 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.924553 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SKGEELFTGV VPILVELDGD VNGHKFSVSG EGEGDATYGK LTLKFICTTG KLPVPWPTLV TTF(CRO)VQCFSR YPDH (MSE)KRHD FFKSA(MSE)PEGY VQERTIFFKD DGNYKTRAEV KFEGDTLVNR IELKGIDFKE DGNILGHKLE YNYNSHN VY I(MSE)ADKQKNGI ...String: SKGEELFTGV VPILVELDGD VNGHKFSVSG EGEGDATYGK LTLKFICTTG KLPVPWPTLV TTF(CRO)VQCFSR YPDH (MSE)KRHD FFKSA(MSE)PEGY VQERTIFFKD DGNYKTRAEV KFEGDTLVNR IELKGIDFKE DGNILGHKLE YNYNSHN VY I(MSE)ADKQKNGI KVNFKIRHNI EDGSVQLADH YQQNTPIGDG PVLLPDNHYL STQSALSKDP NEKRDH(MSE)VLL EFVTAAGI UniProtKB: Green fluorescent protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 6.0 Å Applied symmetry - Helical parameters - Δ&Phi: 138.9 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 5.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 54973 |
|---|---|
| Startup model | Type of model: OTHER / Details: Solid cylinder |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)