+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9015 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Apo-EsCas13d | |||||||||
 Map data Map data | Apo-EsCas13d | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  unidentified bacterium (bacteria) unidentified bacterium (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.5 Å | |||||||||
 Authors Authors | Zhang C / Lyumkis D | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2018 Journal: Cell / Year: 2018Title: Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d. Authors: Cheng Zhang / Silvana Konermann / Nicholas J Brideau / Peter Lotfy / Xuebing Wu / Scott J Novick / Timothy Strutzenberg / Patrick R Griffin / Patrick D Hsu / Dmitry Lyumkis /  Abstract: CRISPR-Cas endonucleases directed against foreign nucleic acids mediate prokaryotic adaptive immunity and have been tailored for broad genetic engineering applications. Type VI-D CRISPR systems ...CRISPR-Cas endonucleases directed against foreign nucleic acids mediate prokaryotic adaptive immunity and have been tailored for broad genetic engineering applications. Type VI-D CRISPR systems contain the smallest known family of single effector Cas enzymes, and their signature Cas13d ribonuclease employs guide RNAs to cleave matching target RNAs. To understand the molecular basis for Cas13d function and explain its compact molecular architecture, we resolved cryoelectron microscopy structures of Cas13d-guide RNA binary complex and Cas13d-guide-target RNA ternary complex to 3.4 and 3.3 Å resolution, respectively. Furthermore, a 6.5 Å reconstruction of apo Cas13d combined with hydrogen-deuterium exchange revealed conformational dynamics that have implications for RNA scanning. These structures, together with biochemical and cellular characterization, provide insights into its RNA-guided, RNA-targeting mechanism and delineate a blueprint for the rational design of improved transcriptome engineering technologies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9015.map.gz emd_9015.map.gz | 56.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9015-v30.xml emd-9015-v30.xml emd-9015.xml emd-9015.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9015.png emd_9015.png | 77.1 KB | ||
| Others |  emd_9015_additional.map.gz emd_9015_additional.map.gz emd_9015_half_map_1.map.gz emd_9015_half_map_1.map.gz emd_9015_half_map_2.map.gz emd_9015_half_map_2.map.gz | 1 MB 11.1 MB 11.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9015 http://ftp.pdbj.org/pub/emdb/structures/EMD-9015 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9015 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9015 | HTTPS FTP |
-Related structure data
| Related structure data |  9013C  9014C  6e9eC  6e9fC C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10229 (Title: Cryo-EM Reconstruction of apo EsCas13d / Data size: 66.9 EMPIAR-10229 (Title: Cryo-EM Reconstruction of apo EsCas13d / Data size: 66.9 Data #1: stack of ~150k particles used for ab initio orientation assignment and reconstruction [picked particles - single frame - unprocessed] Data #2: stack of ~16k best particles from above [picked particles - single frame - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9015.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9015.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Apo-EsCas13d | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.79 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
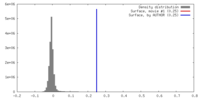
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Apo-EsCas13d, additional map
| File | emd_9015_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Apo-EsCas13d, additional map | ||||||||||||
| Projections & Slices |
| ||||||||||||
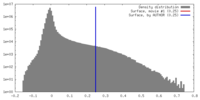
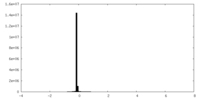
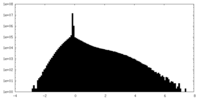
| Density Histograms |
-Half map: Apo-EsCas13d, half map #1
| File | emd_9015_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Apo-EsCas13d, half map #1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
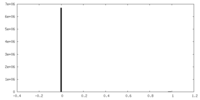
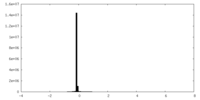
| Density Histograms |
-Half map: Apo-EsCas13d, half map #2
| File | emd_9015_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Apo-EsCas13d, half map #2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
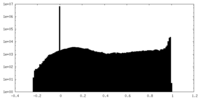
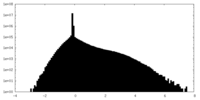
| Density Histograms |
- Sample components
Sample components
-Entire : Apo-EsCas13d
| Entire | Name: Apo-EsCas13d |
|---|---|
| Components |
|
-Supramolecule #1: Apo-EsCas13d
| Supramolecule | Name: Apo-EsCas13d / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  unidentified bacterium (bacteria) unidentified bacterium (bacteria) |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 110 KDa |
-Macromolecule #1: Apo-esCas13d
| Macromolecule | Name: Apo-esCas13d / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  unidentified bacterium (bacteria) unidentified bacterium (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MGKKIHARDL REQRKTDRTE KFADQNKKRE AERAVPKKDA AVSVKSVSSV SSKKDNVTKS MAKAAGVKSV FAVGNTVYMT SFGRGNDAVL EQKIVDTSHE PLNIDDPAYQ LNVVTMNGYS VTGHRGETVS AVTDNPLRRF NGRKKDEPEQ SVPTDMLCLK PTLEKKFFGK ...String: MGKKIHARDL REQRKTDRTE KFADQNKKRE AERAVPKKDA AVSVKSVSSV SSKKDNVTKS MAKAAGVKSV FAVGNTVYMT SFGRGNDAVL EQKIVDTSHE PLNIDDPAYQ LNVVTMNGYS VTGHRGETVS AVTDNPLRRF NGRKKDEPEQ SVPTDMLCLK PTLEKKFFGK EFDDNIHIQL IYNILDIEKI LAVYSTNAIY ALNNMSADEN IENSDFFMKR TTDETFDDFE KKKESTNSRE KADFDAFEKF IGNYRLAYFA DAFYVNKKNP KGKAKNVLRE DKELYSVLTL IGKLRHWCVH SEEGRAEFWL YKLDELKDDF KNVLDVVYNR PVEEINNRFI ENNKVNIQIL GSVYKNTDIA ELVRSYYEFL ITKKYKNMGF SIKKLRESML EGKGYADKEY DSVRNKLYQM TDFILYTGYI NEDSDRADDL VNTLRSSLKE DDKTTVYCKE ADYLWKKYRE SIREVADALD GDNIKKLSKS NIEIQEDKLR KCFISYADSV SEFTKLIYLL TRFLSGKEIN DLVTTLINKF DNIRSFLEIM DELGLDRTFT AEYSFFEGST KYLAELVELN SFVKSCSFDI NAKRTMYRDA LDILGIESDK TEEDIEKMID NILQIDANGD KKLKKNNGLR NFIASNVIDS NRFKYLVRYG NPKKIRETAK CKPAVRFVLN EIPDAQIERY YEACCPKNTA LCSANKRREK LADMIAEIKF ENFSDAGNYQ KANVTSRTSE AEIKRKNQAI IRLYLTVMYI MLKNLVNVNA RYVIAFHCVE RDTKLYAESG LEVGNIEKNK TNLTMAVMGV KLENGIIKTE FDKSFAENAA NRYLRNARWY KLILDNLKKS ERAVVNEFRN TVCHLNAIRN ININIKEIKE VENYFALYHY LIQKHLENRF ADKKVERDTG DFISKLEEHK TYCKDFVKAY CTPFGYNLVR YKNLTIDGLF DKNYPGKDDS DEQK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||||||||
| Grid | Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER / Details: unspecified | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-120 / Number real images: 1158 / Average electron dose: 56.8 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 57000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Target criteria: Correlation coefficient |
|---|
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)