+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9003 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CP506 Fab/BG505 SOSIP.664 complexe | |||||||||
 Map data Map data | CP506 Fab/BG505 SOSIP.664 complexes | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Cavia porcellus (domestic guinea pig) / Cavia porcellus (domestic guinea pig) /   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
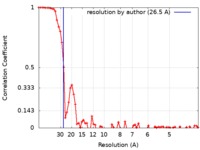
| Method | single particle reconstruction / negative staining / Resolution: 26.5 Å | |||||||||
 Authors Authors | Yang YY / Ward AB | |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2019 Journal: Cell Rep / Year: 2019Title: The HIV-1 Envelope Glycoprotein C3/V4 Region Defines a Prevalent Neutralization Epitope following Immunization. Authors: Lin Lei / Yuhe R Yang / Karen Tran / Yimeng Wang / Chi-I Chiang / Gabriel Ozorowski / Yongli Xiao / Andrew B Ward / Richard T Wyatt / Yuxing Li /  Abstract: Despite recent progress in engineering native trimeric HIV-1 envelope glycoprotein (Env) mimics as vaccine candidates, Env trimers often induce vaccine-matched neutralizing antibody (NAb) responses. ...Despite recent progress in engineering native trimeric HIV-1 envelope glycoprotein (Env) mimics as vaccine candidates, Env trimers often induce vaccine-matched neutralizing antibody (NAb) responses. Understanding the specificities of autologous NAb responses and the underlying molecular mechanisms restricting the neutralization breadth is therefore informative to improve vaccine efficacy. Here, we delineate the response specificity by single B cell sorting and serum analysis of guinea pigs immunized with BG505 SOSIP.664 Env trimers. Our results reveal a prominent immune target containing both conserved and strain-specific residues in the C3/V4 region of Env in trimer-vaccinated animals. The defined NAb response shares a high degree of similarity with the early NAb response developed by a naturally infected infant from whom the HIV virus strain BG505 was isolated and later developed a broadly NAb response. Our study describes strain-specific responses and their possible evolution pathways, thereby highlighting the potential to broaden NAb responses by immunogen re-design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9003.map.gz emd_9003.map.gz | 20.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9003-v30.xml emd-9003-v30.xml emd-9003.xml emd-9003.xml | 10.9 KB 10.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9003_fsc.xml emd_9003_fsc.xml | 8.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_9003.png emd_9003.png | 52.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9003 http://ftp.pdbj.org/pub/emdb/structures/EMD-9003 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9003 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9003 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9003.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9003.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
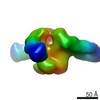
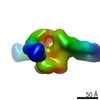
| Annotation | CP506 Fab/BG505 SOSIP.664 complexes | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : CP506 Fab/BG505 SOSIP.664 complex
| Entire | Name: CP506 Fab/BG505 SOSIP.664 complex |
|---|---|
| Components |
|
-Supramolecule #1: CP506 Fab/BG505 SOSIP.664 complex
| Supramolecule | Name: CP506 Fab/BG505 SOSIP.664 complex / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Cavia porcellus (domestic guinea pig) Cavia porcellus (domestic guinea pig) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: BG505 SOSIP.664 Env trimer
| Supramolecule | Name: BG505 SOSIP.664 Env trimer / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: guinea pig CP506 Fab
| Supramolecule | Name: guinea pig CP506 Fab / type: complex / ID: 3 / Parent: 2 |
|---|---|
| Source (natural) | Organism:  Cavia porcellus (domestic guinea pig) Cavia porcellus (domestic guinea pig) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.03 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Staining | Type: NEGATIVE / Material: 2% Uranyl Formate |
| Grid | Model: Homemade / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Average exposure time: 0.5 sec. / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal defocus min: 1.5 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)