[English] 日本語
 Yorodumi
Yorodumi- EMDB-8978: Cryo-EM structure of the active, Gs-protein complexed, human CGRP... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8978 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor | |||||||||
 Map data Map data | Active, Gs-protein complexed, CGRP receptor | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | class B G protein-coupled receptor / agonist-receptor-G protein ternary complex / calcitonin gene-related peptide receptor / receptor activity modifying protein 1 / active-state G protein-coupled receptor / signaling protein / phase plate / phase contrast | |||||||||
| Function / homology |  Function and homology information Function and homology informationnervous system process involved in regulation of systemic arterial blood pressure / calcitonin gene-related peptide binding / : / cellular response to sucrose stimulus / adrenomedullin binding / CGRP receptor complex / adrenomedullin receptor activity / adrenomedullin receptor complex / adrenomedullin receptor signaling pathway / amylin receptor activity ...nervous system process involved in regulation of systemic arterial blood pressure / calcitonin gene-related peptide binding / : / cellular response to sucrose stimulus / adrenomedullin binding / CGRP receptor complex / adrenomedullin receptor activity / adrenomedullin receptor complex / adrenomedullin receptor signaling pathway / amylin receptor activity / calcitonin receptor activity / calcitonin gene-related peptide receptor signaling pathway / vascular associated smooth muscle cell proliferation / calcitonin gene-related peptide receptor activity / positive regulation of interleukin-1 alpha production / negative regulation of calcium ion transport into cytosol / amylin receptor 1 signaling pathway / positive regulation of macrophage differentiation / amylin receptor signaling pathway / Calcitonin-like ligand receptors / regulation of G protein-coupled receptor signaling pathway / vasculature development / G protein-coupled receptor internalization / endothelial cell proliferation / negative regulation of bone resorption / leukocyte cell-cell adhesion / negative regulation of osteoclast differentiation / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / regulation of cytosolic calcium ion concentration / D1 dopamine receptor binding / intracellular transport / endothelial cell migration / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / cellular response to hormone stimulus / coreceptor activity / positive regulation of vascular associated smooth muscle cell proliferation / negative regulation of blood pressure / adenylate cyclase-activating adrenergic receptor signaling pathway / regulation of insulin secretion / cellular response to glucagon stimulus / adenylate cyclase activator activity / trans-Golgi network membrane / positive regulation of interleukin-8 production / protein localization to plasma membrane / intracellular protein transport / negative regulation of inflammatory response to antigenic stimulus / hormone activity / bone development / receptor internalization / G protein-coupled receptor activity / platelet aggregation / regulation of blood pressure / vasodilation / cognition / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / calcium ion transport / sensory perception of smell / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / cell-cell signaling / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / protein transport Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Liang YL / Khoshouei M | |||||||||
| Funding support |  Australia, 2 items Australia, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2018 Journal: Nature / Year: 2018Title: Cryo-EM structure of the active, G-protein complexed, human CGRP receptor. Authors: Yi-Lynn Liang / Maryam Khoshouei / Giuseppe Deganutti / Alisa Glukhova / Cassandra Koole / Thomas S Peat / Mazdak Radjainia / Jürgen M Plitzko / Wolfgang Baumeister / Laurence J Miller / ...Authors: Yi-Lynn Liang / Maryam Khoshouei / Giuseppe Deganutti / Alisa Glukhova / Cassandra Koole / Thomas S Peat / Mazdak Radjainia / Jürgen M Plitzko / Wolfgang Baumeister / Laurence J Miller / Deborah L Hay / Arthur Christopoulos / Christopher A Reynolds / Denise Wootten / Patrick M Sexton /         Abstract: Calcitonin gene-related peptide (CGRP) is a widely expressed neuropeptide that has a major role in sensory neurotransmission. The CGRP receptor is a heterodimer of the calcitonin receptor-like ...Calcitonin gene-related peptide (CGRP) is a widely expressed neuropeptide that has a major role in sensory neurotransmission. The CGRP receptor is a heterodimer of the calcitonin receptor-like receptor (CLR) class B G-protein-coupled receptor and a type 1 transmembrane domain protein, receptor activity-modifying protein 1 (RAMP1). Here we report the structure of the human CGRP receptor in complex with CGRP and the G-protein heterotrimer at 3.3 Å global resolution, determined by Volta phase-plate cryo-electron microscopy. The receptor activity-modifying protein transmembrane domain sits at the interface between transmembrane domains 3, 4 and 5 of CLR, and stabilizes CLR extracellular loop 2. RAMP1 makes only limited direct contact with CGRP, consistent with its function in allosteric modulation of CLR. Molecular dynamics simulations indicate that RAMP1 provides stability to the receptor complex, particularly in the positioning of the extracellular domain of CLR. This work provides insights into the control of G-protein-coupled receptor function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8978.map.gz emd_8978.map.gz | 3.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8978-v30.xml emd-8978-v30.xml emd-8978.xml emd-8978.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8978.png emd_8978.png | 89.6 KB | ||
| Filedesc metadata |  emd-8978.cif.gz emd-8978.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8978 http://ftp.pdbj.org/pub/emdb/structures/EMD-8978 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8978 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8978 | HTTPS FTP |
-Related structure data
| Related structure data |  6e3yMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8978.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8978.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Active, Gs-protein complexed, CGRP receptor | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
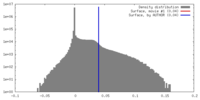
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of a full-length, active-state calcitonin gene-related pe...
| Entire | Name: Complex of a full-length, active-state calcitonin gene-related peptide receptor with calcitonin gene-related peptide ligand, heterotrimeric Gs protein and nanobody35 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of a full-length, active-state calcitonin gene-related pe...
| Supramolecule | Name: Complex of a full-length, active-state calcitonin gene-related peptide receptor with calcitonin gene-related peptide ligand, heterotrimeric Gs protein and nanobody35 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Calcitonin gene-related peptide 1
| Macromolecule | Name: Calcitonin gene-related peptide 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 3.795354 KDa |
| Sequence | String: ACDTATCVTH RLAGLLSRSG GVVKNNFVPT NVGSKAF(NH2) UniProtKB: Calcitonin gene-related peptide 1 |
-Macromolecule #2: Nanobody 35
| Macromolecule | Name: Nanobody 35 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.140742 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGGG LVQPGGSLRL SCAASGFTFS NYKMNWVRQA PGKGLEWVSD ISQSGASISY TGSVKGRFTI SRDNAKNTLY LQMNSLKPE DTAVYYCARC PAPFTRDCFD VTSTTYAYRG QGTQVTVSSH HHHHHEPEA |
-Macromolecule #3: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.683434 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKNTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE ...String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKNTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE YQLIDCAQYF LDKIDVIKQA DYVPSDQDLL RCRVLTSGIF ETKFQVDKVN FHMFDVGAQR DERRKWIQCF ND VTAIIFV VASSSYNMVI REDNQTNRLQ AALKLFDSIW NNKWLRDTSV ILFLNKQDLL AEKVLAGKSK IEDYFPEFAR YTT PEDATP EPGEDPRVTR AKYFIRDEFL RISTASGDGR HYCYPHFTCA VDTENIRRVF NDCRDIIQRM HLRQYELL UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.534062 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MHHHHHHGSS GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC R FLDDNQIV ...String: MHHHHHHGSS GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC R FLDDNQIV TSSGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD IN AICFFPN GNAFATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAG HDNRVS CLGVTDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #5: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #6: Calcitonin gene-related peptide type 1 receptor
| Macromolecule | Name: Calcitonin gene-related peptide type 1 receptor / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 56.27452 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDLEVLFQG PAELEESPED SIQLGVTRNK IMTAQYECYQ KIMQDPIQQA EGVYCNRTWD GWLCWNDVA AGTESMQLCP DYFQDFDPSE KVTKICDQDG NWFRHPASNR TWTNYTQCNV NTHEKVKTAL NLFYLTIIGH G LSIASLLI ...String: MKTIIALSYI FCLVFADYKD DDDLEVLFQG PAELEESPED SIQLGVTRNK IMTAQYECYQ KIMQDPIQQA EGVYCNRTWD GWLCWNDVA AGTESMQLCP DYFQDFDPSE KVTKICDQDG NWFRHPASNR TWTNYTQCNV NTHEKVKTAL NLFYLTIIGH G LSIASLLI SLGIFFYFKS LSCQRITLHK NLFFSFVCNS VVTIIHLTAV ANNQALVATN PVSCKVSQFI HLYLMGCNYF WM LCEGIYL HTLIVVAVFA EKQHLMWYYF LGWGFPLIPA CIHAIARSLY YNDNCWISSD THLLYIIHGP ICAALLVNLF FLL NIVRVL ITKLKVTHQA ESNLYMKAVR ATLILVPLLG IEFVLIPWRP EGKIAEEVYD YIMHILMHFQ GLLVSTIFCF FNGE VQAIL RRNWNQYKIQ FGNSFSNSEA LRSASYTVST ISDGPGYSHD CPSEHLNGKS IHDIENVLLK PENLYNPAGL EVLFQ GPHH HHHHHH UniProtKB: Calcitonin gene-related peptide type 1 receptor |
-Macromolecule #7: Receptor activity-modifying protein 1
| Macromolecule | Name: Receptor activity-modifying protein 1 / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 17.066701 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKHGSCQE ANYGALLREL CLTQFQVDME AVGETLWCDW GRTIRSYREL ADCTWHMAEK LGCFWPNAE VDRFFLAVHG RYFRSCPISG RAVRDPPGSI LYPFIVVPIT VTLLVTALVV WQSKRTEGIV UniProtKB: Receptor activity-modifying protein 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 |
| Vitrification | Cryogen name: NITROGEN / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 47170 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.3 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 407000 |
| Initial angle assignment | Type: COMMON LINE |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)