[English] 日本語
 Yorodumi
Yorodumi- EMDB-8937: Cryo-EM Structure of Mycobacterium smegmatis C(minus) 50S ribosom... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8937 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of Mycobacterium smegmatis C(minus) 50S ribosomal subunit | ||||||||||||
 Map data Map data | Cryo-EM structure of Mycobacterium smegmatis C(minus) 50S ribosomal subunit | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Hibernation factor complex / RIBOSOME | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationlarge ribosomal subunit / transferase activity / ribosomal large subunit assembly / 5S rRNA binding / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / negative regulation of translation / rRNA binding ...large ribosomal subunit / transferase activity / ribosomal large subunit assembly / 5S rRNA binding / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / mRNA binding / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  Mycobacterium smegmatis str. MC2 155 (bacteria) / Mycobacterium smegmatis str. MC2 155 (bacteria) /  Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) | ||||||||||||
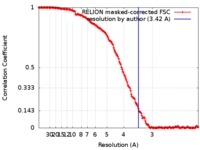
| Method | single particle reconstruction / cryo EM / Resolution: 3.42 Å | ||||||||||||
 Authors Authors | Sharma MR / Li Y / Korripella R / Yang Y / Kaushal PS / Lin Q / Wade JT / Gray AG / Derbyshire KM / Agrawal RK / Ojha A | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2018 Journal: Proc Natl Acad Sci U S A / Year: 2018Title: Zinc depletion induces ribosome hibernation in mycobacteria. Authors: Yunlong Li / Manjuli R Sharma / Ravi K Koripella / Yong Yang / Prem S Kaushal / Qishan Lin / Joseph T Wade / Todd A Gray / Keith M Derbyshire / Rajendra K Agrawal / Anil K Ojha /  Abstract: Bacteria respond to zinc starvation by replacing ribosomal proteins that have the zinc-binding CXXC motif (C+) with their zinc-free (C-) paralogues. Consequences of this process beyond zinc ...Bacteria respond to zinc starvation by replacing ribosomal proteins that have the zinc-binding CXXC motif (C+) with their zinc-free (C-) paralogues. Consequences of this process beyond zinc homeostasis are unknown. Here, we show that the C- ribosome in is the exclusive target of a bacterial protein Y homolog, referred to as mycobacterial-specific protein Y (MPY), which binds to the decoding region of the 30S subunit, thereby inactivating the ribosome. MPY binding is dependent on another mycobacterial protein, MPY recruitment factor (MRF), which is induced on zinc depletion, and interacts with C- ribosomes. MPY binding confers structural stability to C- ribosomes, promoting survival of growth-arrested cells under zinc-limiting conditions. Binding of MPY also has direct influence on the dynamics of aminoglycoside-binding pockets of the C- ribosome to inhibit binding of these antibiotics. Together, our data suggest that zinc limitation leads to ribosome hibernation and aminoglycoside resistance in mycobacteria. Furthermore, our observation of the expression of the proteins of C- ribosomes in in a mouse model of infection suggests that ribosome hibernation could be relevant in our understanding of persistence and drug tolerance of the pathogen encountered during chemotherapy of TB. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8937.map.gz emd_8937.map.gz | 18.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8937-v30.xml emd-8937-v30.xml emd-8937.xml emd-8937.xml | 50.4 KB 50.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8937_fsc.xml emd_8937_fsc.xml | 15.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_8937.png emd_8937.png | 202.3 KB | ||
| Filedesc metadata |  emd-8937.cif.gz emd-8937.cif.gz | 10.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8937 http://ftp.pdbj.org/pub/emdb/structures/EMD-8937 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8937 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8937 | HTTPS FTP |
-Related structure data
| Related structure data |  6dzpMC  8932C  8934C  6dziC  6dzkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8937.map.gz / Format: CCP4 / Size: 357 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8937.map.gz / Format: CCP4 / Size: 357 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of Mycobacterium smegmatis C(minus) 50S ribosomal subunit | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
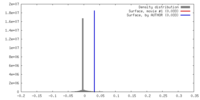
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Mycobacterium smegmatis C(minus) 70S ribosome with MPY
+Supramolecule #1: Mycobacterium smegmatis C(minus) 70S ribosome with MPY
+Macromolecule #1: 23S rRNA
+Macromolecule #2: 5S rRNA
+Macromolecule #3: 50S ribosomal protein L2
+Macromolecule #4: 50S ribosomal protein L3
+Macromolecule #5: 50S ribosomal protein L4
+Macromolecule #6: 50S ribosomal protein L5
+Macromolecule #7: 50S ribosomal protein L6
+Macromolecule #8: 50S ribosomal protein L9
+Macromolecule #9: 50S ribosomal protein L10
+Macromolecule #10: 50S ribosomal protein L11
+Macromolecule #11: 50S ribosomal protein L13
+Macromolecule #12: 50S ribosomal protein L14
+Macromolecule #13: 50S ribosomal protein L15
+Macromolecule #14: 50S ribosomal protein L16
+Macromolecule #15: 50S ribosomal protein L17
+Macromolecule #16: 50S ribosomal protein L18
+Macromolecule #17: 50S ribosomal protein L19
+Macromolecule #18: 50S ribosomal protein L20
+Macromolecule #19: 50S ribosomal protein L21
+Macromolecule #20: 50S ribosomal protein L22
+Macromolecule #21: 50S ribosomal protein L23
+Macromolecule #22: 50S ribosomal protein L24
+Macromolecule #23: 50S ribosomal protein L25
+Macromolecule #24: 50S ribosomal protein L27
+Macromolecule #25: 50S ribosomal protein L29
+Macromolecule #26: 50S ribosomal protein L30
+Macromolecule #27: 50S ribosomal protein L32
+Macromolecule #28: 50S ribosomal protein L33 2
+Macromolecule #29: 50S ribosomal protein L34
+Macromolecule #30: 50S ribosomal protein L35
+Macromolecule #31: 50S ribosomal protein L36
+Macromolecule #32: 50S ribosomal protein L31
+Macromolecule #33: 50S ribosomal protein L28
+Macromolecule #34: Uncharacterized protein
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Support film - Material: CARBON / Support film - topology: CONTINUOUS / Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK I |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 67.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6dzp: |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)