+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8925 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of mouse TRPV3-Y564A | |||||||||
 Map data Map data | primary map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.5 Å | |||||||||
 Authors Authors | Singh AK / McGoldrick LL / Sobolevsky AI | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2018 Journal: Nat Struct Mol Biol / Year: 2018Title: Structure and gating mechanism of the transient receptor potential channel TRPV3. Authors: Appu K Singh / Luke L McGoldrick / Alexander I Sobolevsky /  Abstract: Transient receptor potential vanilloid subfamily member 3 (TRPV3) channel plays a crucial role in skin physiology and pathophysiology. Mutations in TRPV3 are associated with various skin diseases, ...Transient receptor potential vanilloid subfamily member 3 (TRPV3) channel plays a crucial role in skin physiology and pathophysiology. Mutations in TRPV3 are associated with various skin diseases, including Olmsted syndrome, atopic dermatitis, and rosacea. Here we present the cryo-electron microscopy structures of full-length mouse TRPV3 in the closed apo and agonist-bound open states. The agonist binds three allosteric sites distal to the pore. Channel opening is accompanied by conformational changes in both the outer pore and the intracellular gate. The gate is formed by the pore-lining S6 helices that undergo local α-to-π helical transitions, elongate, rotate, and splay apart in the open state. In the closed state, the shorter S6 segments are entirely α-helical, expose their nonpolar surfaces to the pore, and hydrophobically seal the ion permeation pathway. These findings further illuminate TRP channel activation and can aid in the design of drugs for the treatment of inflammatory skin conditions, itch, and pain. | |||||||||
| History |
|
- Structure visualization
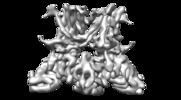
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8925.map.gz emd_8925.map.gz | 8.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8925-v30.xml emd-8925-v30.xml emd-8925.xml emd-8925.xml | 10.2 KB 10.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8925.png emd_8925.png | 98.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8925 http://ftp.pdbj.org/pub/emdb/structures/EMD-8925 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8925 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8925 | HTTPS FTP |
-Related structure data
| Related structure data |  8919C  8920C  8921C  6dvwC  6dvyC  6dvzC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8925.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8925.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | primary map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.98 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : TRPV3
| Entire | Name: TRPV3 |
|---|---|
| Components |
|
-Supramolecule #1: TRPV3
| Supramolecule | Name: TRPV3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: TRPV3-Y564A
| Macromolecule | Name: TRPV3-Y564A / type: protein_or_peptide / ID: 1 / Enantiomer: DEXTRO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: mgahskemap lmgkrttapg gnpvvltekr padltptkks ahffleiegf epnptvtkts ppifskpmds nirqclsgnc ddmdspqspq ddvtetpsnp nspsanlake eqrqkkkrlk krifaavseg cveelrellq dlqdlcrrrr gldvpdflmh kltasdtgkt ...String: mgahskemap lmgkrttapg gnpvvltekr padltptkks ahffleiegf epnptvtkts ppifskpmds nirqclsgnc ddmdspqspq ddvtetpsnp nspsanlake eqrqkkkrlk krifaavseg cveelrellq dlqdlcrrrr gldvpdflmh kltasdtgkt clmkallnin pntkeivril lafaeendil drfinaeyte eayegqtaln iaierrqgdi tavliaagad vnahakgvff npkyqhegfy fgetplalaa ctnqpeivql lmeneqtdit sqdsrgnnil halvtvaedf ktqndfvkrm ydmillrsgn weletmrnnd gltplqlaak mgkaeilkyi lsreikekpl rslsrkftdw aygpvsssly dltnvdtttd nsvleiivyn tnidnrheml tleplhtllh tkwkkfakym fflsfcfyff ynitltlvsy yrprededlp hplalthkms wlqllgrmfv liwatcisvk egiaifllrp sdlqsilsda wfhfvffvqa vlvilsvfly lfaykeylac lvlamalgwa nmlaytrgfq smgmysvmiq kvilhdvlkf lfvyilfllg fgvalaslie kcskdkkdcs sygsfsdavl elfkltiglg dlniqqnsty pilflfllit yviltfvlll nmlialmget venvskeser iwrlqrarti lefekmlpew lrsrfrmgel ckvadedfrl clrinevkwt ewkthvsfln edpgpirrta dlnkiqdssr snskttlyaf deldefpets v |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.0 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Material: GOLD / Support film - Material: CARBON / Support film - topology: HOLEY | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 67.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 6.5 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 44661 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















