[English] 日本語
 Yorodumi
Yorodumi- EMDB-8791: MERS S ectodomain trimer in complex with Fab of neutralizing anti... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8791 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | MERS S ectodomain trimer in complex with Fab of neutralizing antibody G4 | |||||||||
 Map data Map data | MERS S ectodomain trimer in complex with variable domain of neutralizing antibody G4 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Immunogen / peplomer / viral spike / MERS / MERS S protein / antibody / G4 / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmembrane fusion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
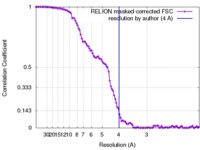
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Pallesen J / Ward AB | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2017 Journal: Proc Natl Acad Sci U S A / Year: 2017Title: Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Authors: Jesper Pallesen / Nianshuang Wang / Kizzmekia S Corbett / Daniel Wrapp / Robert N Kirchdoerfer / Hannah L Turner / Christopher A Cottrell / Michelle M Becker / Lingshu Wang / Wei Shi / Wing- ...Authors: Jesper Pallesen / Nianshuang Wang / Kizzmekia S Corbett / Daniel Wrapp / Robert N Kirchdoerfer / Hannah L Turner / Christopher A Cottrell / Michelle M Becker / Lingshu Wang / Wei Shi / Wing-Pui Kong / Erica L Andres / Arminja N Kettenbach / Mark R Denison / James D Chappell / Barney S Graham / Andrew B Ward / Jason S McLellan /  Abstract: Middle East respiratory syndrome coronavirus (MERS-CoV) is a lineage C betacoronavirus that since its emergence in 2012 has caused outbreaks in human populations with case-fatality rates of ∼36%. ...Middle East respiratory syndrome coronavirus (MERS-CoV) is a lineage C betacoronavirus that since its emergence in 2012 has caused outbreaks in human populations with case-fatality rates of ∼36%. As in other coronaviruses, the spike (S) glycoprotein of MERS-CoV mediates receptor recognition and membrane fusion and is the primary target of the humoral immune response during infection. Here we use structure-based design to develop a generalizable strategy for retaining coronavirus S proteins in the antigenically optimal prefusion conformation and demonstrate that our engineered immunogen is able to elicit high neutralizing antibody titers against MERS-CoV. We also determined high-resolution structures of the trimeric MERS-CoV S ectodomain in complex with G4, a stem-directed neutralizing antibody. The structures reveal that G4 recognizes a glycosylated loop that is variable among coronaviruses and they define four conformational states of the trimer wherein each receptor-binding domain is either tightly packed at the membrane-distal apex or rotated into a receptor-accessible conformation. Our studies suggest a potential mechanism for fusion initiation through sequential receptor-binding events and provide a foundation for the structure-based design of coronavirus vaccines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8791.map.gz emd_8791.map.gz | 95.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8791-v30.xml emd-8791-v30.xml emd-8791.xml emd-8791.xml | 24.1 KB 24.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8791_fsc.xml emd_8791_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_8791.png emd_8791.png | 82.9 KB | ||
| Filedesc metadata |  emd-8791.cif.gz emd-8791.cif.gz | 7.1 KB | ||
| Others |  emd_8791_half_map_1.map.gz emd_8791_half_map_1.map.gz emd_8791_half_map_2.map.gz emd_8791_half_map_2.map.gz | 84.1 MB 84.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8791 http://ftp.pdbj.org/pub/emdb/structures/EMD-8791 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8791 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8791 | HTTPS FTP |
-Related structure data
| Related structure data |  5w9pMC  8783C  8784C  8785C  8786C  8787C  8788C  8789C  8790C  8792C  8793C  5vyhC  5vzrC  5w9hC  5w9iC  5w9jC  5w9kC  5w9lC  5w9mC  5w9nC  5w9oC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8791.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8791.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MERS S ectodomain trimer in complex with variable domain of neutralizing antibody G4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.02 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
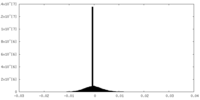
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: MERS S ectodomain trimer in complex with variable...
| File | emd_8791_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MERS S ectodomain trimer in complex with variable domain of neutralizing antibody G4 | ||||||||||||
| Projections & Slices |
| ||||||||||||
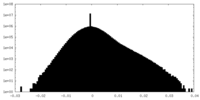
| Density Histograms |
-Half map: MERS S ectodomain trimer in complex with variable...
| File | emd_8791_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MERS S ectodomain trimer in complex with variable domain of neutralizing antibody G4 | ||||||||||||
| Projections & Slices |
| ||||||||||||
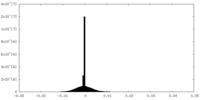
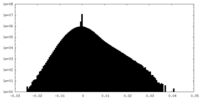
| Density Histograms |
- Sample components
Sample components
-Entire : MERS S ectodomain trimer in complex with Fab of neutralizing anti...
| Entire | Name: MERS S ectodomain trimer in complex with Fab of neutralizing antibody G4 |
|---|---|
| Components |
|
-Supramolecule #1: MERS S ectodomain trimer in complex with Fab of neutralizing anti...
| Supramolecule | Name: MERS S ectodomain trimer in complex with Fab of neutralizing antibody G4 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 600 KDa |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 146.325969 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MIHSVFLLMF LLTPTESYVD VGPDSVKSAC IEVDIQQTFF DKTWPRPIDV SKADGIIYPQ GRTYSNITIT YQGLFPYQGD HGDMYVYSA GHATGTTPQK LFVANYSQDV KQFANGFVVR IGAAANSTGT VIISPSTSAT IRKIYPAFML GSSVGNFSDG K MGRFFNHT ...String: MIHSVFLLMF LLTPTESYVD VGPDSVKSAC IEVDIQQTFF DKTWPRPIDV SKADGIIYPQ GRTYSNITIT YQGLFPYQGD HGDMYVYSA GHATGTTPQK LFVANYSQDV KQFANGFVVR IGAAANSTGT VIISPSTSAT IRKIYPAFML GSSVGNFSDG K MGRFFNHT LVLLPDGCGT LLRAFYCILE PRSGNHCPAG NSYTSFATYH TPATDCSDGN YNRNASLNSF KEYFNLRNCT FM YTYNITE DEILEWFGIT QTAQGVHLFS SRYVDLYGGN MFQFATLPVY DTIKYYSIIP HSIRSIQSDR KAWAAFYVYK LQP LTFLLD FSVDGYIRRA IDCGFNDLSQ LHCSYESFDV ESGVYSVSSF EAKPSGSVVE QAEGVECDFS PLLSGTPPQV YNFK RLVFT NCNYNLTKLL SLFSVNDFTC SQISPAAIAS NCYSSLILDY FSYPLSMKSD LSVSSAGPIS QFNYKQSFSN PTCLI LATV PHNLTTITKP LKYSYINKCS RFLSDDRTEV PQLVNANQYS PCVSIVPSTV WEDGDYYRKQ LSPLEGGGWL VASGST VAM TEQLQMGFGI TVQYGTDTNS VCPKLEFAND TKIASQLGNC VEYSLYGVSG RGVFQNCTAV GVRQQRFVYD AYQNLVG YY SDDGNYYCLR ACVSVPVSVI YDKETKTHAT LFGSVACEHI SSTMSQYSRS TRSMLKRRDS TYGPLQTPVG CVLGLVNS S LFVEDCKLPL GQSLCALPDT PSTLTPASVG SVPGEMRLAS IAFNHPIQVD QLNSSYFKLS IPTNFSFGVT QEYIQTTIQ KVTVDCKQYV CNGFQKCEQL LREYGQFCSK INQALHGANL RQDDSVRNLF ASVKSSQSSP IIPGFGGDFN LTLLEPVSIS TGSRSARSA IEDLLFDKVT IADPGYMQGY DDCMQQGPAS ARDLICAQYV AGYKVLPPLM DVNMEAAYTS SLLGSIAGVG W TAGLSSFA AIPFAQSIFY RLNGVGITQQ VLSENQKLIA NKFNQALGAM QTGFTTTNEA FHKVQDAVNN NAQALSKLAS EL SNTFGAI SASIGDIIQR LDPPEQDAQI DRLINGRLTT LNAFVAQQLV RSESAALSAQ LAKDKVNECV KAQSKRSGFC GQG THIVSF VVNAPNGLYF MHVGYYPSNH IEVVSAYGLC DAANPTNCIA PVNGYFIKTN NTRIVDEWSY TGSSFYAPEP ITSL NTKYV APQVTYQNIS TNLPPPLLGN STGIDFQDEL DEFFKNVSTS IPNFGSLTQI NTTLLDLTYE MLSLQQVVKA LNESY IDLK ELGNYTYGSG YIPEAPRDGQ AYVRKDGEWV LLSTFLGRSL EVLFQ UniProtKB: Spike glycoprotein |
-Macromolecule #2: G4 VH
| Macromolecule | Name: G4 VH / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.312352 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLQQSGPE LVRPGVSVKI SCKGSGYTFT DYAIHWVKQS HAKSLEWIGV FSTYYGNTNY NQKFKGRATM TVDKSSSTAY MELARLTSE DSAIYYCARK SYYVDYVDAM DYWGQGTSVT VSSASTTPPS VYPLAPGSAA QTNSMVTLGC LVKGYFPEPV T VTWNSGSL ...String: QVQLQQSGPE LVRPGVSVKI SCKGSGYTFT DYAIHWVKQS HAKSLEWIGV FSTYYGNTNY NQKFKGRATM TVDKSSSTAY MELARLTSE DSAIYYCARK SYYVDYVDAM DYWGQGTSVT VSSASTTPPS VYPLAPGSAA QTNSMVTLGC LVKGYFPEPV T VTWNSGSL SSGVHTFPAV LQSDLYTLSS SVTVPSSTWP SETVTCNVAH PASSTKVDKK IVPRDCGKGL EVLFQ |
-Macromolecule #3: G4 VL
| Macromolecule | Name: G4 VL / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.867367 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVLTQSPAS LAVSLGQRAT ISCRASESVD NYGISFMNWF QQKPGQPPKL LISATSNQGS GVPARFIGSG SGTDFSLNIH PVEEDDTAM YFCQQSKEVP RTFGGGTKLE IKRTDAAPTV SIFPPSSEQL TSGGASVVCF LNNFYPKDIN VKWKIDGSER Q NGVLNSWT ...String: DIVLTQSPAS LAVSLGQRAT ISCRASESVD NYGISFMNWF QQKPGQPPKL LISATSNQGS GVPARFIGSG SGTDFSLNIH PVEEDDTAM YFCQQSKEVP RTFGGGTKLE IKRTDAAPTV SIFPPSSEQL TSGGASVVCF LNNFYPKDIN VKWKIDGSER Q NGVLNSWT DQDSKDSTYS MSSTLTLTKD EYERHNSYTC EATHKTSTSP IVKSFNRNEC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Name: TBS |
| Grid | Model: C-flat / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 5 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-35 / Number grids imaged: 1 / Average exposure time: 0.2 sec. / Average electron dose: 1.89 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-5w9p: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)