[English] 日本語
 Yorodumi
Yorodumi- EMDB-8578: Negative Stain EM of C05 mutant Fab (VPGSGW) and CR9114 Fab in co... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8578 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
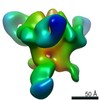
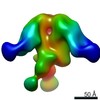
| Title | Negative Stain EM of C05 mutant Fab (VPGSGW) and CR9114 Fab in complex with H1 HA Trimer | |||||||||
 Map data Map data | EM negative stain map of CR9114 and VPGSGW fabs in complex with H1 hemagglutinin trimer. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationclathrin-dependent endocytosis of virus by host cell / apical plasma membrane / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane Similarity search - Function | |||||||||
| Biological species |  Influenza A virus (A/Solomon Islands/3/2006(H1N1)) Influenza A virus (A/Solomon Islands/3/2006(H1N1)) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 18.0 Å | |||||||||
 Authors Authors | Turner HL / Ward AB | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2017 Journal: Nat Commun / Year: 2017Title: In vitro evolution of an influenza broadly neutralizing antibody is modulated by hemagglutinin receptor specificity. Authors: Nicholas C Wu / Geramie Grande / Hannah L Turner / Andrew B Ward / Jia Xie / Richard A Lerner / Ian A Wilson /  Abstract: The relatively recent discovery and characterization of human broadly neutralizing antibodies (bnAbs) against influenza virus provide valuable insights into antiviral and vaccine development. ...The relatively recent discovery and characterization of human broadly neutralizing antibodies (bnAbs) against influenza virus provide valuable insights into antiviral and vaccine development. However, the factors that influence the evolution of high-affinity bnAbs remain elusive. We therefore explore the functional sequence space of bnAb C05, which targets the receptor-binding site (RBS) of influenza haemagglutinin (HA) via a long CDR H3. We combine saturation mutagenesis with yeast display to enrich for C05 variants of CDR H3 that bind to H1 and H3 HAs. The C05 variants evolve up to 20-fold higher affinity but increase specificity to each HA subtype used in the selection. Structural analysis reveals that the fine specificity is strongly influenced by a highly conserved substitution that regulates receptor binding in different subtypes. Overall, this study suggests that subtle natural variations in the HA RBS between subtypes and species may differentially influence the evolution of high-affinity bnAbs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8578.map.gz emd_8578.map.gz | 10.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8578-v30.xml emd-8578-v30.xml emd-8578.xml emd-8578.xml | 9.2 KB 9.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8578.png emd_8578.png | 33.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8578 http://ftp.pdbj.org/pub/emdb/structures/EMD-8578 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8578 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8578 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8578.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8578.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM negative stain map of CR9114 and VPGSGW fabs in complex with H1 hemagglutinin trimer. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Negative stain EM map of C05 Mutant Fab (VPGSGW) and CR9114 fabs ...
| Entire | Name: Negative stain EM map of C05 Mutant Fab (VPGSGW) and CR9114 fabs in complex with H1 HA trimer. |
|---|---|
| Components |
|
-Supramolecule #1: Negative stain EM map of C05 Mutant Fab (VPGSGW) and CR9114 fabs ...
| Supramolecule | Name: Negative stain EM map of C05 Mutant Fab (VPGSGW) and CR9114 fabs in complex with H1 HA trimer. type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Solomon Islands/3/2006(H1N1)) Influenza A virus (A/Solomon Islands/3/2006(H1N1))Strain: A/Solomon Islands/3/2006(H1N1) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.019 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Name: Tris Buffer Saline / Details: Solution was made fresh using 0.2um filter. |
| Staining | Type: NEGATIVE / Material: Uranyl Formate Details: 0.2% Uranyl Formate. Samples were placed on 400 mesh copper grids covered with nitrocellulose. Samples were blotted off and UF was added for a total of 30 seconds before being blotted off. |
| Grid | Model: EMS / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number real images: 93 / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 18874 |
|---|---|
| Final reconstruction | Number classes used: 1 / Applied symmetry - Point group: C1 (asymmetric) / Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 18.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: sxali3d.py / Number images used: 18874 |
| Initial angle assignment | Type: COMMON LINE |
| Final angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: SPARX |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















