+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8589 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram average of microtubules from Trypanosoma brucei | |||||||||
 Map data Map data | Subtomogram average of microtubules from Trypanosoma brucei | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 55.0 Å | |||||||||
 Authors Authors | Chen M / Sun SY / Ludtke SJ / He CY / Schmid MF / Chiu W | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Methods / Year: 2017 Journal: Nat Methods / Year: 2017Title: Convolutional neural networks for automated annotation of cellular cryo-electron tomograms. Authors: Muyuan Chen / Wei Dai / Stella Y Sun / Darius Jonasch / Cynthia Y He / Michael F Schmid / Wah Chiu / Steven J Ludtke /   Abstract: Cellular electron cryotomography offers researchers the ability to observe macromolecules frozen in action in situ, but a primary challenge with this technique is identifying molecular components ...Cellular electron cryotomography offers researchers the ability to observe macromolecules frozen in action in situ, but a primary challenge with this technique is identifying molecular components within the crowded cellular environment. We introduce a method that uses neural networks to dramatically reduce the time and human effort required for subcellular annotation and feature extraction. Subsequent subtomogram classification and averaging yield in situ structures of molecular components of interest. The method is available in the EMAN2.2 software package. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8589.map.gz emd_8589.map.gz | 25 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8589-v30.xml emd-8589-v30.xml emd-8589.xml emd-8589.xml | 9.1 KB 9.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8589.png emd_8589.png | 52.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8589 http://ftp.pdbj.org/pub/emdb/structures/EMD-8589 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8589 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8589 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8589.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8589.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram average of microtubules from Trypanosoma brucei | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
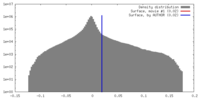
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Microtubule
| Entire | Name: Microtubule |
|---|---|
| Components |
|
-Supramolecule #1: Microtubule
| Supramolecule | Name: Microtubule / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2200FS |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-12 (4k x 3k) / Detector mode: INTEGRATING / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 55.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: EMAN (ver. 2.2) / Number subtomograms used: 511 |
|---|---|
| Extraction | Number tomograms: 1 / Number images used: 511 / Reference model: Reference free averaging / Method: Automated tomogram segmentation / Software - Name: EMAN (ver. 2.2) / Software - details: Tomoseg tool |
| Final angle assignment | Type: OTHER / Software - Name: EMAN (ver. 2.2) / Details: Single particle tomography tool in EMAN2 |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)