[English] 日本語
 Yorodumi
Yorodumi- EMDB-8516: Structure of human cystic fibrosis transmembrane conductance regu... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8516 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human cystic fibrosis transmembrane conductance regulator (CFTR) | |||||||||
 Map data Map data | Human cystic fibrosis transmembrane conductance regulator, B sharpened map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of voltage-gated chloride channel activity / : / Sec61 translocon complex binding / channel-conductance-controlling ATPase / intracellularly ATP-gated chloride channel activity / positive regulation of enamel mineralization / transepithelial water transport / RHO GTPases regulate CFTR trafficking / amelogenesis / intracellular pH elevation ...positive regulation of voltage-gated chloride channel activity / : / Sec61 translocon complex binding / channel-conductance-controlling ATPase / intracellularly ATP-gated chloride channel activity / positive regulation of enamel mineralization / transepithelial water transport / RHO GTPases regulate CFTR trafficking / amelogenesis / intracellular pH elevation / chloride channel inhibitor activity / : / multicellular organismal-level water homeostasis / chloride channel regulator activity / Golgi-associated vesicle membrane / cholesterol transport / bicarbonate transport / bicarbonate transmembrane transporter activity / membrane hyperpolarization / vesicle docking involved in exocytosis / chloride transmembrane transporter activity / cholesterol biosynthetic process / sperm capacitation / RHOQ GTPase cycle / chloride channel activity / positive regulation of exocytosis / ATPase-coupled transmembrane transporter activity / ABC-type transporter activity / chloride channel complex / positive regulation of insulin secretion involved in cellular response to glucose stimulus / 14-3-3 protein binding / cellular response to forskolin / chloride transmembrane transport / cellular response to cAMP / response to endoplasmic reticulum stress / PDZ domain binding / establishment of localization in cell / clathrin-coated endocytic vesicle membrane / Defective CFTR causes cystic fibrosis / Late endosomal microautophagy / recycling endosome / ABC-family proteins mediated transport / transmembrane transport / recycling endosome membrane / Chaperone Mediated Autophagy / Aggrephagy / Cargo recognition for clathrin-mediated endocytosis / protein-folding chaperone binding / Clathrin-mediated endocytosis / early endosome membrane / early endosome / apical plasma membrane / endosome membrane / Ub-specific processing proteases / lysosomal membrane / endoplasmic reticulum membrane / enzyme binding / cell surface / ATP hydrolysis activity / protein-containing complex / ATP binding / membrane / nucleus / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.87 Å | |||||||||
 Authors Authors | Liu F / Zhang Z / Chen J | |||||||||
 Citation Citation |  Journal: Cell / Year: 2016 Journal: Cell / Year: 2016Title: Atomic Structure of the Cystic Fibrosis Transmembrane Conductance Regulator. Authors: Zhe Zhang / Jue Chen /  Abstract: The cystic fibrosis transmembrane conductance regulator (CFTR) is an anion channel evolved from the ATP-binding cassette (ABC) transporter family. In this study, we determined the structure of ...The cystic fibrosis transmembrane conductance regulator (CFTR) is an anion channel evolved from the ATP-binding cassette (ABC) transporter family. In this study, we determined the structure of zebrafish CFTR in the absence of ATP by electron cryo-microscopy to 3.7 Å resolution. Human and zebrafish CFTR share 55% sequence identity, and 42 of the 46 cystic-fibrosis-causing missense mutational sites are identical. In CFTR, we observe a large anion conduction pathway lined by numerous positively charged residues. A single gate near the extracellular surface closes the channel. The regulatory domain, dephosphorylated, is located in the intracellular opening between the two nucleotide-binding domains (NBDs), preventing NBD dimerization and channel opening. The structure also reveals why many cystic-fibrosis-causing mutations would lead to defects either in folding, ion conduction, or gating and suggests new avenues for therapeutic intervention. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8516.map.gz emd_8516.map.gz | 200.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8516-v30.xml emd-8516-v30.xml emd-8516.xml emd-8516.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8516.png emd_8516.png | 139.1 KB | ||
| Others |  emd_8516_additional.map.gz emd_8516_additional.map.gz | 18.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8516 http://ftp.pdbj.org/pub/emdb/structures/EMD-8516 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8516 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8516 | HTTPS FTP |
-Related structure data
| Related structure data |  5uakM  8461C  5uarC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8516.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8516.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human cystic fibrosis transmembrane conductance regulator, B sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.817 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
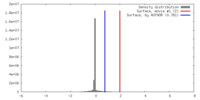
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Human cystic fibrosis transmembrane conductance regulator without B...
| File | emd_8516_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human cystic fibrosis transmembrane conductance regulator without B factor sharpening (FREALIGN) | ||||||||||||
| Projections & Slices |
| ||||||||||||
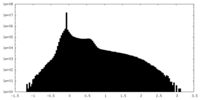
| Density Histograms |
- Sample components
Sample components
-Entire : Cystic fibrosis transmembrane conductance regulator or ABCC7
| Entire | Name: Cystic fibrosis transmembrane conductance regulator or ABCC7 |
|---|---|
| Components |
|
-Supramolecule #1: Cystic fibrosis transmembrane conductance regulator or ABCC7
| Supramolecule | Name: Cystic fibrosis transmembrane conductance regulator or ABCC7 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK 293S GnTI- / Recombinant plasmid: pEG Bacmam Homo sapiens (human) / Recombinant cell: HEK 293S GnTI- / Recombinant plasmid: pEG Bacmam |
| Molecular weight | Experimental: 168 kDa/nm |
-Macromolecule #1: Cystic fibrosis transmembrane conductance regulator
| Macromolecule | Name: Cystic fibrosis transmembrane conductance regulator / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: ec: 3.6.3.49 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 169.353578 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MQRSPLEKAS VVSKLFFSWT RPILRKGYRQ RLELSDIYQI PSVDSADNLS EKLEREWDRE LASKKNPKLI NALRRCFFWR FMFYGIFLY LGEVTKAVQP LLLGRIIASY DPDNKEERSI AIYLGIGLCL LFIVRTLLLH PAIFGLHHIG MQMRIAMFSL I YKKTLKLS ...String: MQRSPLEKAS VVSKLFFSWT RPILRKGYRQ RLELSDIYQI PSVDSADNLS EKLEREWDRE LASKKNPKLI NALRRCFFWR FMFYGIFLY LGEVTKAVQP LLLGRIIASY DPDNKEERSI AIYLGIGLCL LFIVRTLLLH PAIFGLHHIG MQMRIAMFSL I YKKTLKLS SRVLDKISIG QLVSLLSNNL NKFDEGLALA HFVWIAPLQV ALLMGLIWEL LQASAFCGLG FLIVLALFQA GL GRMMMKY RDQRAGKISE RLVITSEMIE NIQSVKAYCW EEAMEKMIEN LRQTELKLTR KAAYVRYFNS SAFFFSGFFV VFL SVLPYA LIKGIILRKI FTTISFCIVL RMAVTRQFPW AVQTWYDSLG AINKIQDFLQ KQEYKTLEYN LTTTEVVMEN VTAF WEEGF GELFEKAKQN NNNRKTSNGD DSLFFSNFSL LGTPVLKDIN FKIERGQLLA VAGSTGAGKT SLLMVIMGEL EPSEG KIKH SGRISFCSQF SWIMPGTIKE NIIFGVSYDE YRYRSVIKAC QLEEDISKFA EKDNIVLGEG GITLSGGQRA RISLAR AVY KDADLYLLDS PFGYLDVLTE KEIFESCVCK LMANKTRILV TSKMEHLKKA DKILILHEGS SYFYGTFSEL QNLQPDF SS KLMGCDSFDQ FSAERRNSIL TETLHRFSLE GDAPVSWTET KKQSFKQTGE FGEKRKNSIL NPINSIRKFS IVQKTPLQ M NGIEEDSDEP LERRLSLVPD SEQGEAILPR ISVISTGPTL QARRRQSVLN LMTHSVNQGQ NIHRKTTAST RKVSLAPQA NLTELDIYSR RLSQETGLEI SEEINEEDLK ECFFDDMESI PAVTTWNTYL RYITVHKSLI FVLIWCLVIF LAEVAASLVV LWLLGNTPL QDKGNSTHSR NNSYAVIITS TSSYYVFYIY VGVADTLLAM GFFRGLPLVH TLITVSKILH HKMLHSVLQA P MSTLNTLK AGGILNRFSK DIAILDDLLP LTIFDFIQLL LIVIGAIAVV AVLQPYIFVA TVPVIVAFIM LRAYFLQTSQ QL KQLESEG RSPIFTHLVT SLKGLWTLRA FGRQPYFETL FHKALNLHTA NWFLYLSTLR WFQMRIEMIF VIFFIAVTFI SIL TTGEGE GRVGIILTLA MNIMSTLQWA VNSSIDVDSL MRSVSRVFKF IDMPTEGKPT KSTKPYKNGQ LSKVMIIENS HVKK DDIWP SGGQMTVKDL TAKYTEGGNA ILENISFSIS PGQRVGLLGR TGSGKSTLLS AFLRLLNTEG EIQIDGVSWD SITLQ QWRK AFGVIPQKVF IFSGTFRKNL DPYEQWSDQE IWKVADEVGL RSVIEQFPGK LDFVLVDGGC VLSHGHKQLM CLARSV LSK AKILLLDEPS AHLDPVTYQI IRRTLKQAFA DCTVILCEHR IEAMLECQQF LVIEENKVRQ YDSIQKLLNE RSLFRQA IS PSDRVKLFPH RNSSKCKSKP QIAALKEETE EEVQDTRLSN SLEVLFQ |
-Macromolecule #2: Cystic fibrosis transmembrane conductance regulator
| Macromolecule | Name: Cystic fibrosis transmembrane conductance regulator / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.549902 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK I |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-50 / Number grids imaged: 1 / Number real images: 3642 / Average exposure time: 0.14 sec. / Average electron dose: 1.68 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 61200 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)