+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8466 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Myeloma IgG4 Structure | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 32.0 Å | |||||||||
 Authors Authors | Ryazantsev S / Tischenko V / Zavyalov V | |||||||||
 Citation Citation |  Journal: Mol Immunol / Year: 2017 Journal: Mol Immunol / Year: 2017Title: Human myeloma IgG4 reveals relatively rigid asymmetric Y-like structure with different conformational stability of C2 domains. Authors: Vladimir M Tischenko / Vladimir P Zav'yalov / Sergey N Ryazantsev /    Abstract: Human IgG4 (hIgG4) has weak pro-inflammatory activity. The structural basis for this is still unclear. Here a 3D model of myeloma hIgG4 was created at ∼3nm resolution using electron microscopy (EM) ...Human IgG4 (hIgG4) has weak pro-inflammatory activity. The structural basis for this is still unclear. Here a 3D model of myeloma hIgG4 was created at ∼3nm resolution using electron microscopy (EM) with negative staining and single-particle 3D reconstruction. The hIgG4 model reveals relatively rigid asymmetric Y-like structure. The model shows that one Fab subunit is closer to the upper portion of the Fc subunit (C2 domain) than the other Fab. This is in agreement with X-ray crystallography and X-ray/neutron scattering, recently published by others. The same hIgG4 sample was studied with differential scanning calorimetry (DSC) and fluorescence. The thermodynamics and fluorescence observations indicate that one C2 domain displays less conformational stability than the other. This finding is consistent with the flipping of one C2 domain, observed in pembrolizumab (recombinant hIgG4) by X-ray crystallography. The specific feature of hIgG4 C2 domains together with relatively rigid asymmetric Y-like structure, in which one Fab subunit is closer to the upper portion of the Fc subunit (C2 domain) than the other Fab, can explain the unique biological properties of hIgG4, such as its weak pro-inflammatory activity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8466.map.gz emd_8466.map.gz | 23.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8466-v30.xml emd-8466-v30.xml emd-8466.xml emd-8466.xml | 10.4 KB 10.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8466.png emd_8466.png emd_8466_1.png emd_8466_1.png emd_8466_2.png emd_8466_2.png | 170.2 KB 170.2 KB 41.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8466 http://ftp.pdbj.org/pub/emdb/structures/EMD-8466 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8466 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8466 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8466.map.gz / Format: CCP4 / Size: 28.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8466.map.gz / Format: CCP4 / Size: 28.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : human myeloma IgG4
| Entire | Name: human myeloma IgG4 |
|---|---|
| Components |
|
-Supramolecule #1: human myeloma IgG4
| Supramolecule | Name: human myeloma IgG4 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Details: Isolated from the serum and purified to 96% purity |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Tissue: blood/serum Homo sapiens (human) / Tissue: blood/serum |
| Molecular weight | Experimental: 160 KDa |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 7.8 / Component - Concentration: 20.0 mM / Component - Name: ammonium acetate |
| Staining | Type: NEGATIVE / Material: 1% uranyl acetate Details: Negative staining with uranyl acetate was carried out using the approach described by Valentine et al. with modification. |
| Grid | Model: Homemade / Material: COPPER / Mesh: 150 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Details | Human myeloma IgG4 |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 100CX |
|---|---|
| Details | images were close to focus |
| Image recording | Film or detector model: KODAK 4489 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Average exposure time: 1.5 sec. / Average electron dose: 100.0 e/Å2 |
| Electron beam | Acceleration voltage: 80 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Calibrated magnification: 80000 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 1.9 mm |
| Sample stage | Specimen holder model: JEOL |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera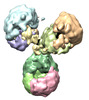







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















