[English] 日本語
 Yorodumi
Yorodumi- EMDB-8349: Cryo-EM Structure of Yeast Pre-40S, Tsr1 region local class "T2" -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8349 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of Yeast Pre-40S, Tsr1 region local class "T2" | ||||||||||||
 Map data Map data | Pre-40S, Tsr1 local class "T2" | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  | ||||||||||||
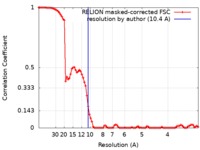
| Method | single particle reconstruction / cryo EM / Resolution: 10.4 Å | ||||||||||||
 Authors Authors | Johnson MC / Ghalei H / Doxtader KA / Karbstein K / Stroupe ME | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Structure / Year: 2017 Journal: Structure / Year: 2017Title: Structural Heterogeneity in Pre-40S Ribosomes. Authors: Matthew C Johnson / Homa Ghalei / Katelyn A Doxtader / Katrin Karbstein / M Elizabeth Stroupe /  Abstract: Late-stage 40S ribosome assembly is a highly regulated dynamic process that occurs in the cytoplasm, alongside the full translation machinery. Seven assembly factors (AFs) regulate and facilitate ...Late-stage 40S ribosome assembly is a highly regulated dynamic process that occurs in the cytoplasm, alongside the full translation machinery. Seven assembly factors (AFs) regulate and facilitate maturation, but the mechanisms through which they work remain undetermined. Here, we present a series of structures of the immature small subunit (pre-40S) determined by three-dimensional (3D) cryoelectron microscopy with 3D sorting to assess the molecule's heterogeneity. These structures demonstrate an extensive structural heterogeneity of interface AFs that likely regulates subunit joining during 40S maturation. We also present structural models for the beak and the platform, two regions where the low resolution of previous studies did not allow for localization of AFs and the rRNA, respectively. These models are supported by biochemical analyses using point variants and suggest that maturation of the 18S 3' end is regulated by dissociation of the AF Dim1 from the subunit interface, consistent with previous biochemical analyses. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8349.map.gz emd_8349.map.gz | 2.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8349-v30.xml emd-8349-v30.xml emd-8349.xml emd-8349.xml | 9.9 KB 9.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8349_fsc.xml emd_8349_fsc.xml | 8.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_8349.png emd_8349.png | 63.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8349 http://ftp.pdbj.org/pub/emdb/structures/EMD-8349 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8349 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8349 | HTTPS FTP |
-Related structure data
| Related structure data |  8346C  8347C  8348C  8350C  8351C  8352C  8353C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8349.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8349.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Pre-40S, Tsr1 local class "T2" | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.62 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : ribosomal small subunit late-stage assembly intermediate
| Entire | Name: ribosomal small subunit late-stage assembly intermediate |
|---|---|
| Components |
|
-Supramolecule #1: ribosomal small subunit late-stage assembly intermediate
| Supramolecule | Name: ribosomal small subunit late-stage assembly intermediate type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)