[English] 日本語
 Yorodumi
Yorodumi- EMDB-8321: Near-atomic resolution cryo-EM reconstruction of doubly bound Tax... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8321 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Near-atomic resolution cryo-EM reconstruction of doubly bound Taxol- and peloruside-stabilized microtubule | ||||||||||||
 Map data Map data | Peloruside- and taxol-bound microtubules | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Taxol / peloruside / microtubule / STRUCTURAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationMicrotubule-dependent trafficking of connexons from Golgi to the plasma membrane / Resolution of Sister Chromatid Cohesion / Hedgehog 'off' state / Cilium Assembly / Intraflagellar transport / COPI-dependent Golgi-to-ER retrograde traffic / Mitotic Prometaphase / Carboxyterminal post-translational modifications of tubulin / RHOH GTPase cycle / EML4 and NUDC in mitotic spindle formation ...Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane / Resolution of Sister Chromatid Cohesion / Hedgehog 'off' state / Cilium Assembly / Intraflagellar transport / COPI-dependent Golgi-to-ER retrograde traffic / Mitotic Prometaphase / Carboxyterminal post-translational modifications of tubulin / RHOH GTPase cycle / EML4 and NUDC in mitotic spindle formation / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins / PKR-mediated signaling / Separation of Sister Chromatids / The role of GTSE1 in G2/M progression after G2 checkpoint / Aggrephagy / RHO GTPases activate IQGAPs / RHO GTPases Activate Formins / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / MHC class II antigen presentation / Recruitment of NuMA to mitotic centrosomes / COPI-mediated anterograde transport / microtubule-based process / structural constituent of cytoskeleton / microtubule cytoskeleton organization / neuron migration / mitotic cell cycle / microtubule cytoskeleton / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / microtubule / hydrolase activity / GTPase activity / GTP binding / metal ion binding / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.5 Å | ||||||||||||
 Authors Authors | Kellogg EH / Nogales E | ||||||||||||
| Funding support |  United States, United States,  Spain, 3 items Spain, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2017 Journal: J Mol Biol / Year: 2017Title: Insights into the Distinct Mechanisms of Action of Taxane and Non-Taxane Microtubule Stabilizers from Cryo-EM Structures. Authors: Elizabeth H Kellogg / Nisreen M A Hejab / Stuart Howes / Peter Northcote / John H Miller / J Fernando Díaz / Kenneth H Downing / Eva Nogales /    Abstract: A number of microtubule (MT)-stabilizing agents (MSAs) have demonstrated or predicted potential as anticancer agents, but a detailed structural basis for their mechanism of action is still lacking. ...A number of microtubule (MT)-stabilizing agents (MSAs) have demonstrated or predicted potential as anticancer agents, but a detailed structural basis for their mechanism of action is still lacking. We have obtained high-resolution (3.9-4.2Å) cryo-electron microscopy (cryo-EM) reconstructions of MTs stabilized by the taxane-site binders Taxol and zampanolide, and by peloruside, which targets a distinct, non-taxoid pocket on β-tubulin. We find that each molecule has unique distinct structural effects on the MT lattice structure. Peloruside acts primarily at lateral contacts and has an effect on the "seam" of heterologous interactions, enforcing a conformation more similar to that of homologous (i.e., non-seam) contacts by which it regularizes the MT lattice. In contrast, binding of either Taxol or zampanolide induces MT heterogeneity. In doubly bound MTs, peloruside overrides the heterogeneity induced by Taxol binding. Our structural analysis illustrates distinct mechanisms of these drugs for stabilizing the MT lattice and is of relevance to the possible use of combinations of MSAs to regulate MT activity and improve therapeutic potential. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8321.map.gz emd_8321.map.gz | 482.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8321-v30.xml emd-8321-v30.xml emd-8321.xml emd-8321.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8321.png emd_8321.png | 452.4 KB | ||
| Filedesc metadata |  emd-8321.cif.gz emd-8321.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8321 http://ftp.pdbj.org/pub/emdb/structures/EMD-8321 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8321 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8321 | HTTPS FTP |
-Related structure data
| Related structure data |  5syeMC  8320C  8322C  8323C  5sycC  5syfC  5sygC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8321.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8321.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Peloruside- and taxol-bound microtubules | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.32 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
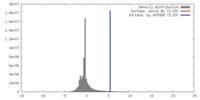
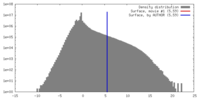
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ternary complex of Taxol- and peloruside-stabilized microtubule l...
| Entire | Name: Ternary complex of Taxol- and peloruside-stabilized microtubule lattice, alpha-tubulin, and beta-tubulin |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of Taxol- and peloruside-stabilized microtubule l...
| Supramolecule | Name: Ternary complex of Taxol- and peloruside-stabilized microtubule lattice, alpha-tubulin, and beta-tubulin type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Tubulin alpha chain
| Macromolecule | Name: Tubulin alpha chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.679051 KDa |
| Sequence | String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIIDLVL DRIRKLADQC TGLQGFLVFH SFGGGTGSGF TSLLMERLSV D YGKKSKLE ...String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIIDLVL DRIRKLADQC TGLQGFLVFH SFGGGTGSGF TSLLMERLSV D YGKKSKLE FSIYPAPQVS TAVVEPYNSI LTTHTTLEHS DCAFMVDNEA IYDICRRNLD IERPTYTNLN RLISQIVSSI TA SLRFDGA LNVDLTEFQT NLVPYPRIHF PLATYAPVIS AEKAYHEQLS VAEITNACFE PANQMVKCDP RHGKYMACCL LYR GDVVPK DVNAAIATIK TKRTIQFVDW CPTGFKVGIN YQPPTVVPGG DLAKVQRAVC MLSNTTAIAE AWARLDHKFD LMYA KRAFV HWYVGEGMEE GEFSEAREDM AALEKDYEEV GV UniProtKB: Tubulin alpha chain |
-Macromolecule #2: Tubulin beta chain
| Macromolecule | Name: Tubulin beta chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 47.825859 KDa |
| Sequence | String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEAAGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKESESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTFS ...String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEAAGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKESESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTFS VVPSPKVSDT VVEPYNATLS VHQLVENTDE TYCIDNEALY DICFRTLKLT TPTYGDLNHL VSATMSGVTT CL RFPGQLN ADLRKLAVNM VPFPRLHFFM PGFAPLTSRG SQQYRALTVP ELTQQMFDAK NMMAACDPRH GRYLTVAAVF RGR MSMKEV DEQMLNVQNK NSSYFVEWIP NNVKTAVCDI PPRGLKMSAT FIGNSTAIQE LFKRISEQFT AMFRRKAFLH WYTG EGMDE MEFTEAESNM NDLVSEYQQY Q UniProtKB: Tubulin beta chain |
-Macromolecule #3: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 1 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Macromolecule #6: TAXOL
| Macromolecule | Name: TAXOL / type: ligand / ID: 6 / Number of copies: 1 / Formula: TA1 |
|---|---|
| Molecular weight | Theoretical: 853.906 Da |
| Chemical component information |  ChemComp-TA1: |
-Macromolecule #7: Peloruside A
| Macromolecule | Name: Peloruside A / type: ligand / ID: 7 / Number of copies: 1 / Formula: POU |
|---|---|
| Molecular weight | Theoretical: 548.663 Da |
| Chemical component information | 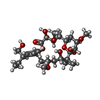 ChemComp-POU: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.8 Component:
| ||||||||||||||||||
| Grid | Model: Protochips CF-1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 20 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: Blot for 4 seconds, blot force 10, before plunging into liquid ethane (FEI VITROBOT MARK IV).. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Temperature | Min: 93.0 K / Max: 93.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-20 / Number grids imaged: 1 / Number real images: 312 / Average exposure time: 0.3 sec. / Average electron dose: 8.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 3.8000000000000003 µm / Calibrated defocus min: 1.1 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 27500 |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 9.43 Å Applied symmetry - Helical parameters - Δ&Phi: -27.7 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Algorithm: FOURIER SPACE / Resolution.type: BY AUTHOR / Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: FREALIGN (ver. 9.09) / Number images used: 17069 |
|---|---|
| Segment selection | Number selected: 27275 Details: Microtubule filaments were manually selected using manualpicker.py. |
| Startup model | Type of model: OTHER Details: Low-pass filtered reference model of microtubules decorated with kinesin |
| Final angle assignment | Type: NOT APPLICABLE / Software - Name: FREALIGN (ver. 9.09) |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: correlation coefficient |
|---|---|
| Output model |  PDB-5sye: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)