+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8288 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Putative tetramer of human CPAP (residues 897-1338) | |||||||||
 Map data Map data | Contour level of 0.031 using Chimera | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 23.0 Å | |||||||||
 Authors Authors | Alvarez-Cabrera AL / Delgado S / Gil D / Mortuza G / Montoya G / Sorzano CO / Tang TK / Carazo JM | |||||||||
 Citation Citation |  Journal: Front Mol Biosci / Year: 2017 Journal: Front Mol Biosci / Year: 2017Title: Electron Microscopy Structural Insights into CPAP Oligomeric Behavior: A Plausible Assembly Process of a Supramolecular Scaffold of the Centrosome. Authors: Ana L Alvarez-Cabrera / Sandra Delgado / David Gil-Carton / Gulnahar B Mortuza / Guillermo Montoya / Carlos O S Sorzano / Tang K Tang / Jose M Carazo /     Abstract: Centrosomal P4.1-associated protein (CPAP) is a cell cycle regulated protein fundamental for centrosome assembly and centriole elongation. In humans, the region between residues 897-1338 of CPAP ...Centrosomal P4.1-associated protein (CPAP) is a cell cycle regulated protein fundamental for centrosome assembly and centriole elongation. In humans, the region between residues 897-1338 of CPAP mediates interactions with other proteins and includes a homodimerization domain. CPAP mutations cause primary autosomal recessive microcephaly and Seckel syndrome. Despite of the biological/clinical relevance of CPAP, its mechanistic behavior remains unclear and its C-terminus (the G-box/TCP domain) is the only part whose structure has been solved. This situation is perhaps due in part to the challenges that represent obtaining the protein in a soluble, homogeneous state for structural studies. Our work constitutes a systematic structural analysis on multiple oligomers of , using single-particle electron microscopy (EM) of negatively stained (NS) samples. Based on image classification into clearly different regular 3D maps (putatively corresponding to dimers and tetramers) and direct observation of individual images representing other complexes of CPAP (i.e., putative flexible monomers and higher-order multimers), we report a dynamic oligomeric behavior of this protein, where different homo-oligomers coexist in variable proportions. We propose that dimerization of the putative homodimer forms a putative tetramer which could be the structural unit for the scaffold that either tethers the pericentriolar material to centrioles or promotes procentriole elongation. A coarse fitting of atomic models into the NS 3D maps at resolutions around 20 Å is performed only to complement our experimental data, allowing us to hypothesize on the oligomeric composition of the different complexes. In this way, the current EM work represents an initial step toward the structural characterization of different oligomers of CPAP, suggesting further insights to understand how this protein works, contributing to the elucidation of control mechanisms for centriole biogenesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8288.map.gz emd_8288.map.gz | 126.4 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8288-v30.xml emd-8288-v30.xml emd-8288.xml emd-8288.xml | 12.3 KB 12.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8288.png emd_8288.png | 32.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8288 http://ftp.pdbj.org/pub/emdb/structures/EMD-8288 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8288 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8288 | HTTPS FTP |
-Validation report
| Summary document |  emd_8288_validation.pdf.gz emd_8288_validation.pdf.gz | 78.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8288_full_validation.pdf.gz emd_8288_full_validation.pdf.gz | 77.5 KB | Display | |
| Data in XML |  emd_8288_validation.xml.gz emd_8288_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8288 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8288 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8288 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8288 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8288.map.gz / Format: CCP4 / Size: 844.7 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8288.map.gz / Format: CCP4 / Size: 844.7 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Contour level of 0.031 using Chimera | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.42 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
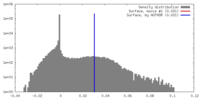
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Putative homotetramer of human CPAP (residues 897-1338)
| Entire | Name: Putative homotetramer of human CPAP (residues 897-1338) |
|---|---|
| Components |
|
-Supramolecule #1: Putative homotetramer of human CPAP (residues 897-1338)
| Supramolecule | Name: Putative homotetramer of human CPAP (residues 897-1338) type: complex / ID: 1 / Parent: 0 Details: Human CPAP (residues 897-1338) expresed in E. coli and purified by Immobilized Metal Affinity Chromatography (IMAC) and Size Exclusion Chromatography (SEC) |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Recombinant plasmid: pST66Trc2-His (is a pET3a modified plasmid) |
| Molecular weight | Experimental: 208 KDa |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
Details: TCEP added to the buffer was prepared fresh | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Formate (0.75%) | |||||||||||||||
| Grid | Model: Quantifoil. Formvar/Carbon / Material: COPPER / Mesh: 400 / Support film - #0 - Film type ID: 1 / Support film - #0 - Material: CARBON / Support film - #0 - topology: CONTINUOUS / Support film - #1 - Film type ID: 2 / Support film - #1 - Material: FORMVAR / Support film - #1 - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Details | This human CPAP construct includes residues 897-1338 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (4k x 4k) / Number grids imaged: 1 / Average exposure time: 1.0 sec. / Average electron dose: 9.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.26 mm |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera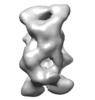







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)