+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-8239 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | EM map of the intact yeast Elongator complex | |||||||||
 マップデータ マップデータ | Endogenously purified yeast Elongator complex | |||||||||
 試料 試料 |
| |||||||||
| 生物種 |  | |||||||||
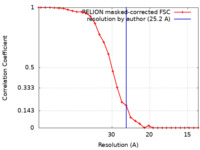
| 手法 | 単粒子再構成法 / ネガティブ染色法 / 解像度: 25.2 Å | |||||||||
 データ登録者 データ登録者 | Setiaputra D / Cheng DTH / Hansen JM / Yip CK | |||||||||
| 資金援助 |  カナダ, 1件 カナダ, 1件
| |||||||||
 引用 引用 |  ジャーナル: EMBO Rep / 年: 2017 ジャーナル: EMBO Rep / 年: 2017タイトル: Molecular architecture of the yeast Elongator complex reveals an unexpected asymmetric subunit arrangement. 著者: Dheva T Setiaputra / Derrick Th Cheng / Shan Lu / Jesse M Hansen / Udit Dalwadi / Cindy Hy Lam / Jeffrey L To / Meng-Qiu Dong / Calvin K Yip /   要旨: Elongator is a ~850 kDa protein complex involved in multiple processes from transcription to tRNA modification. Conserved from yeast to humans, Elongator is assembled from two copies of six unique ...Elongator is a ~850 kDa protein complex involved in multiple processes from transcription to tRNA modification. Conserved from yeast to humans, Elongator is assembled from two copies of six unique subunits (Elp1 to Elp6). Despite the wealth of structural data on the individual subunits, the overall architecture and subunit organization of the full Elongator and the molecular mechanisms of how it exerts its multiple activities remain unclear. Using single-particle electron microscopy (EM), we revealed that yeast Elongator adopts a bilobal architecture and an unexpected asymmetric subunit arrangement resulting from the hexameric Elp456 subassembly anchored to one of the two Elp123 lobes that form the structural scaffold. By integrating the EM data with available subunit crystal structures and restraints generated from cross-linking coupled to mass spectrometry, we constructed a multiscale molecular model that showed the two Elp3, the main catalytic subunit, are located in two distinct environments. This work provides the first structural insights into Elongator and a framework to understand the molecular basis of its multifunctionality. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_8239.map.gz emd_8239.map.gz | 714.3 KB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-8239-v30.xml emd-8239-v30.xml emd-8239.xml emd-8239.xml | 13.9 KB 13.9 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_8239_fsc.xml emd_8239_fsc.xml | 2.6 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_8239.png emd_8239.png | 74.6 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8239 http://ftp.pdbj.org/pub/emdb/structures/EMD-8239 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8239 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8239 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_8239_validation.pdf.gz emd_8239_validation.pdf.gz | 427.7 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_8239_full_validation.pdf.gz emd_8239_full_validation.pdf.gz | 427.2 KB | 表示 | |
| XML形式データ |  emd_8239_validation.xml.gz emd_8239_validation.xml.gz | 6.1 KB | 表示 | |
| CIF形式データ |  emd_8239_validation.cif.gz emd_8239_validation.cif.gz | 7.6 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8239 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8239 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8239 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8239 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_8239.map.gz / 形式: CCP4 / 大きさ: 1.4 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_8239.map.gz / 形式: CCP4 / 大きさ: 1.4 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Endogenously purified yeast Elongator complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : Intact yeast Elongator complex
| 全体 | 名称: Intact yeast Elongator complex |
|---|---|
| 要素 |
|
-超分子 #1: Intact yeast Elongator complex
| 超分子 | 名称: Intact yeast Elongator complex / タイプ: complex / ID: 1 / 親要素: 0 / 詳細: Endogenously purified yeast Elongator complex |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 850 KDa |
-実験情報
-構造解析
| 手法 | ネガティブ染色法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.4 構成要素:
詳細: The final purification buffer was part of a glycerol gradient containing crosslinker as per the GraFix technique (Stark 2010): 40 mM HEPES, pH 7.4, 150 mM NaCl, 0.1% Tween-20, 1 mM EDTA, ~20% ...詳細: The final purification buffer was part of a glycerol gradient containing crosslinker as per the GraFix technique (Stark 2010): 40 mM HEPES, pH 7.4, 150 mM NaCl, 0.1% Tween-20, 1 mM EDTA, ~20% glycerol, ~0.02% glutaraldehyde. Fractions containing the sample were concentrated and the buffer was exchanged for the final buffer: 40 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM EDTA. | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 染色 | タイプ: NEGATIVE / 材質: Uranyl formate 詳細: Sample was applied to a glow-discharged copper grid overlaid with carbon (carbon evaporator and amyl acetate) and stained with uranyl formate. | ||||||||||||
| グリッド | モデル: Ted Pella Gilder Grids / 材質: COPPER / メッシュ: 400 / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: CONTINUOUS / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 時間: 25 sec. / 前処理 - 雰囲気: AIR / 前処理 - 気圧: 0.039 kPa |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TECNAI SPIRIT |
|---|---|
| 撮影 | フィルム・検出器のモデル: FEI EAGLE (4k x 4k) / デジタル化 - サイズ - 横: 4096 pixel / デジタル化 - サイズ - 縦: 4096 pixel / 撮影したグリッド数: 1 / 実像数: 100 / 平均露光時間: 1.0 sec. / 平均電子線量: 25.0 e/Å2 |
| 電子線 | 加速電圧: 120 kV / 電子線源: LAB6 |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 倍率(公称値): 49000 |
| 試料ステージ | 試料ホルダーモデル: SIDE ENTRY, EUCENTRIC |
| 実験機器 |  モデル: Tecnai Spirit / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー



 UCSF Chimera
UCSF Chimera








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)