[English] 日本語
 Yorodumi
Yorodumi- EMDB-8193: Cryo-EM structure of isocitrate dehydrogenase (IDH1) in complex w... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8193 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of isocitrate dehydrogenase (IDH1) in complex with ML309 inhibitor | |||||||||
 Map data Map data | Isocitrate dehydrogenase (IDH1) in complex with ML309 inhibitor | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | isocitrate dehydrogenase / small metabolic complex / small molecule inhibitor / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationAbnormal conversion of 2-oxoglutarate to 2-hydroxyglutarate / NADPH regeneration / regulation of phospholipid catabolic process / regulation of phospholipid biosynthetic process / NFE2L2 regulating TCA cycle genes / isocitrate metabolic process / isocitrate dehydrogenase (NADP+) / isocitrate dehydrogenase (NADP+) activity / NADPH regeneration / NADP+ metabolic process ...Abnormal conversion of 2-oxoglutarate to 2-hydroxyglutarate / NADPH regeneration / regulation of phospholipid catabolic process / regulation of phospholipid biosynthetic process / NFE2L2 regulating TCA cycle genes / isocitrate metabolic process / isocitrate dehydrogenase (NADP+) / isocitrate dehydrogenase (NADP+) activity / NADPH regeneration / NADP+ metabolic process / 2-oxoglutarate metabolic process / glyoxylate cycle / response to steroid hormone / female gonad development / peroxisomal matrix / tricarboxylic acid cycle / glutathione metabolic process / Peroxisomal protein import / NAD binding / tertiary granule lumen / peroxisome / NADP binding / response to oxidative stress / secretory granule lumen / ficolin-1-rich granule lumen / cadherin binding / Neutrophil degranulation / magnesium ion binding / protein homodimerization activity / mitochondrion / extracellular exosome / extracellular region / identical protein binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Merk A / Bartesaghi A | |||||||||
 Citation Citation |  Journal: Cell / Year: 2016 Journal: Cell / Year: 2016Title: Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery. Authors: Alan Merk / Alberto Bartesaghi / Soojay Banerjee / Veronica Falconieri / Prashant Rao / Mindy I Davis / Rajan Pragani / Matthew B Boxer / Lesley A Earl / Jacqueline L S Milne / Sriram Subramaniam /  Abstract: Recent advances in single-particle cryoelecton microscopy (cryo-EM) are enabling generation of numerous near-atomic resolution structures for well-ordered protein complexes with sizes ≥ ∼200 kDa. ...Recent advances in single-particle cryoelecton microscopy (cryo-EM) are enabling generation of numerous near-atomic resolution structures for well-ordered protein complexes with sizes ≥ ∼200 kDa. Whether cryo-EM methods are equally useful for high-resolution structural analysis of smaller, dynamic protein complexes such as those involved in cellular metabolism remains an important question. Here, we present 3.8 Å resolution cryo-EM structures of the cancer target isocitrate dehydrogenase (93 kDa) and identify the nature of conformational changes induced by binding of the allosteric small-molecule inhibitor ML309. We also report 2.8-Å- and 1.8-Å-resolution structures of lactate dehydrogenase (145 kDa) and glutamate dehydrogenase (334 kDa), respectively. With these results, two perceived barriers in single-particle cryo-EM are overcome: (1) crossing 2 Å resolution and (2) obtaining structures of proteins with sizes < 100 kDa, demonstrating that cryo-EM can be used to investigate a broad spectrum of drug-target interactions and dynamic conformational states. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8193.map.gz emd_8193.map.gz | 26.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8193-v30.xml emd-8193-v30.xml emd-8193.xml emd-8193.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8193.png emd_8193.png | 52.8 KB | ||
| Filedesc metadata |  emd-8193.cif.gz emd-8193.cif.gz | 6.2 KB | ||
| Others |  emd_8193_additional_1.map.gz emd_8193_additional_1.map.gz emd_8193_additional_2.map.gz emd_8193_additional_2.map.gz | 27.2 MB 27.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8193 http://ftp.pdbj.org/pub/emdb/structures/EMD-8193 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8193 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8193 | HTTPS FTP |
-Related structure data
| Related structure data |  5k11MC  8191C  8192C  8194C  5k0zC  5k10C  5k12C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8193.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8193.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Isocitrate dehydrogenase (IDH1) in complex with ML309 inhibitor | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.495 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
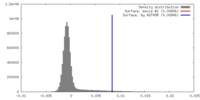
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Map sharpened using a B-factor of -180
| File | emd_8193_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map sharpened using a B-factor of -180 | ||||||||||||
| Projections & Slices |
| ||||||||||||
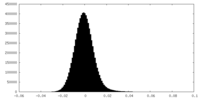
| Density Histograms |
-Additional map: Reconstruction obtained without imposing symmetry
| File | emd_8193_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction obtained without imposing symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
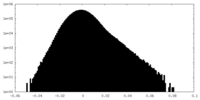
| Density Histograms |
- Sample components
Sample components
-Entire : Isocitrate dehydrogenase R132C mutant in complex with ML309
| Entire | Name: Isocitrate dehydrogenase R132C mutant in complex with ML309 |
|---|---|
| Components |
|
-Supramolecule #1: Isocitrate dehydrogenase R132C mutant in complex with ML309
| Supramolecule | Name: Isocitrate dehydrogenase R132C mutant in complex with ML309 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 93 KDa |
-Macromolecule #1: Isocitrate dehydrogenase [NADP] cytoplasmic
| Macromolecule | Name: Isocitrate dehydrogenase [NADP] cytoplasmic / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: isocitrate dehydrogenase (NADP+) |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 46.334742 KDa |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: KKISGGSVVE MQGDEMTRII WELIKEKLIF PYVELDLHSY DLGIENRDAT NDQVTKDAAE AIKKHNVGVK CATITPDEKR VEEFKLKQM WKSPNGTIRN ILGGTVFREA IICKNIPRLV SGWVKPIIIG CHAYGDQYRA TDFVVPGPGK VEITYTPSDG T QKVTYLVH ...String: KKISGGSVVE MQGDEMTRII WELIKEKLIF PYVELDLHSY DLGIENRDAT NDQVTKDAAE AIKKHNVGVK CATITPDEKR VEEFKLKQM WKSPNGTIRN ILGGTVFREA IICKNIPRLV SGWVKPIIIG CHAYGDQYRA TDFVVPGPGK VEITYTPSDG T QKVTYLVH NFEEGGGVAM GMYNQDKSIE DFAHSSFQMA LSKGWPLYLS TKNTILKKYD GRFKDIFQEI YDKQYKSQFE AQ KIWYEHR LIDDMVAQAM KSEGGFIWAC KNYDGDVQSD SVAQGYGSLG MMTSVLVCPD GKTVEAEAAH GTVTRHYRMY QKG QETSTN PIASIFAWTR GLAHRAKLDN NKELAFFANA LEEVSIETIE AGFMTKDLAA CIKGLPNVQR SDYLNTFEFM DKLG ENLKI KLAQAK UniProtKB: Isocitrate dehydrogenase [NADP] cytoplasmic |
-Macromolecule #2: NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE
| Macromolecule | Name: NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE type: ligand / ID: 2 / Number of copies: 2 / Formula: NDP |
|---|---|
| Molecular weight | Theoretical: 745.421 Da |
| Chemical component information |  ChemComp-NDP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.8 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM GP / Details: Plunged into liquid ethane (LEICA EM GP). |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 79.6 K / Max: 79.8 K |
| Specialist optics | Energy filter - Name: GIF Quantum / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 0-29 / Number real images: 820 / Average exposure time: 0.2 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 101000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 270000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-5k11: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)