[English] 日本語
 Yorodumi
Yorodumi- EMDB-8140: 3.87 Angstrom structure of the nucleosome core particle obtained ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8140 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 3.87 Angstrom structure of the nucleosome core particle obtained by phase-plate cryo-EM | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
| Biological species | ||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.87 Å | |||||||||
 Authors Authors | Chua EYD / Vogirala VK / Inian O / Wong ASW / Plitzko JM / Danev R / Sandin S | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2016 Journal: Nucleic Acids Res / Year: 2016Title: 3.9 Å structure of the nucleosome core particle determined by phase-plate cryo-EM. Authors: Eugene Y D Chua / Vinod K Vogirala / Oviya Inian / Andrew S W Wong / Lars Nordenskiöld / Juergen M Plitzko / Radostin Danev / Sara Sandin /   Abstract: The Volta phase plate is a recently developed electron cryo-microscopy (cryo-EM) device that enables contrast enhancement of biological samples. Here we have evaluated the potential of combining ...The Volta phase plate is a recently developed electron cryo-microscopy (cryo-EM) device that enables contrast enhancement of biological samples. Here we have evaluated the potential of combining phase-plate imaging and single particle analysis to determine the structure of a small protein-DNA complex. To test the method, we made use of a 200 kDa Nucleosome Core Particle (NCP) reconstituted with 601 DNA for which a high-resolution X-ray crystal structure is known. We find that the phase plate provides a significant contrast enhancement that permits individual NCPs and DNA to be clearly identified in amorphous ice. The refined structure from 26,060 particles has an overall resolution of 3.9 Å and the density map exhibits structural features consistent with the estimated resolution, including clear density for amino acid side chains and DNA features such as the phosphate backbone. Our results demonstrate that phase-plate cryo-EM promises to become an important method to determine novel near-atomic resolution structures of small and challenging samples, such as nucleosomes in complex with nucleosome-binding factors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8140.map.gz emd_8140.map.gz | 14.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8140-v30.xml emd-8140-v30.xml emd-8140.xml emd-8140.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
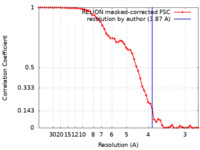
| FSC (resolution estimation) |  emd_8140_fsc.xml emd_8140_fsc.xml | 5.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_8140.png emd_8140.png | 21.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8140 http://ftp.pdbj.org/pub/emdb/structures/EMD-8140 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8140 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8140 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8140.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8140.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.38 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Nucleosome core particle with 145 base pairs of the Widom 601 seq...
| Entire | Name: Nucleosome core particle with 145 base pairs of the Widom 601 sequence. |
|---|---|
| Components |
|
-Supramolecule #1: Nucleosome core particle with 145 base pairs of the Widom 601 seq...
| Supramolecule | Name: Nucleosome core particle with 145 base pairs of the Widom 601 sequence. type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Experimental: 200 KDa |
-Supramolecule #2: Widom 601 DNA, 145 base pairs
| Supramolecule | Name: Widom 601 DNA, 145 base pairs / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Experimental: 90 KDa |
-Supramolecule #3: Histone octamer
| Supramolecule | Name: Histone octamer / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#5 |
|---|---|
| Source (natural) | Organism: |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 110 KDa |
-Macromolecule #1: Widom 601 DNA, 145 base pairs
| Macromolecule | Name: Widom 601 DNA, 145 base pairs / type: dna / ID: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Sequence | String: ATCAGAATCC CGGTGCCGAG GCCGCTCAAT TGGTCGTAGA CAGCTCTAGC ACCGCTTAAA CGCACGTACG CGCTGTCCCC CGCGTTTTAA CCGCCAAGGG GATTACTCCC TAGTCTCCAG GCACGTGTCA GATATATACA TCGAT |
-Macromolecule #2: Histone 2A
| Macromolecule | Name: Histone 2A / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Recombinant expression | Organism:  |
| Sequence | String: SGRGKQGGKT RAKAKTRSSR AGLQFPVGRV HRLLRKGNYA ERVGAGAPVY LAAVLEYLTA EILELAGNAA RDNKKTRIIP RHLQLAVRND EELNKLLGRV TIAQGGVLPN IQSVLLPKKT ESSKSKSK |
-Macromolecule #3: Histone 2B
| Macromolecule | Name: Histone 2B / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Recombinant expression | Organism:  |
| Sequence | String: PEPAKSAPAP KKGSKKAVTK TQKKDGKKRR KTRKESYAIY VYKVLKQVHP DTGISSKAMS IMNSFVNDVF ERIAGEASRL AHYNKRSTIT SREIQTAVRL LLPGELAKHA VSEGTKAVTK YTSAK |
-Macromolecule #4: Histone 3
| Macromolecule | Name: Histone 3 / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Recombinant expression | Organism:  |
| Sequence | String: ARTKQTARKS TGGKAPRKQL ATKAARKSAP ATGGVKKPHR YRPGTVALRE IRRYQKSTEL LIRKLPFQRL VREIAQDFKT DLRFQSSAVM ALQEASEAYL VALFEDTNLC AIHAKRVTIM PKDIQLARRI RGERA |
-Macromolecule #5: Histone 4
| Macromolecule | Name: Histone 4 / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Recombinant expression | Organism:  |
| Sequence | String: SGRGKGGKGL GKGGAKRHRK VLRDNIQGIT KPAIRRLARR GGVKRISGLI YEETRGVLKV FLENVIRDAV TYTEHAKRKT VTAMDVVYAL KRQGRTLYGF GG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/2 or R1/4 / Material: COPPER/RHODIUM / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK III / Details: 3-4 seconds blotting time. | ||||||||||||
| Details | This sample was monodispersed |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-20 / Average exposure time: 10.0 sec. / Average electron dose: 31.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 78.96 |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)