[English] 日本語
 Yorodumi
Yorodumi- EMDB-8130: Architecture of ovine respiratory supercomplex (I-III2-IV) confor... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8130 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Architecture of ovine respiratory supercomplex (I-III2-IV) conformation tight | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information: / : / regulation of oxidative phosphorylation / respiratory chain complex IV assembly / mitochondrial respirasome assembly / : / respiratory chain complex IV / cytochrome-c oxidase / respiratory chain complex III / oxidative phosphorylation ...: / : / regulation of oxidative phosphorylation / respiratory chain complex IV assembly / mitochondrial respirasome assembly / : / respiratory chain complex IV / cytochrome-c oxidase / respiratory chain complex III / oxidative phosphorylation / quinol-cytochrome-c reductase / mitochondrial electron transport, cytochrome c to oxygen / quinol-cytochrome-c reductase activity / cytochrome-c oxidase activity / mitochondrial electron transport, ubiquinol to cytochrome c / electron transport coupled proton transport / membrane => GO:0016020 / ATP synthesis coupled electron transport / metalloendopeptidase activity / 2 iron, 2 sulfur cluster binding / electron transfer activity / oxidoreductase activity / mitochondrial inner membrane / copper ion binding / heme binding / ubiquitin protein ligase binding / proteolysis / nucleoplasm / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
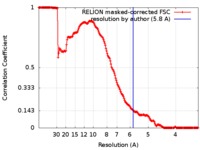
| Method | single particle reconstruction / cryo EM / Resolution: 5.8 Å | |||||||||
 Authors Authors | Sazanov LA / Letts JA / Fiedorczuk K | |||||||||
 Citation Citation |  Journal: Nature / Year: 2016 Journal: Nature / Year: 2016Title: The architecture of respiratory supercomplexes. Authors: James A Letts / Karol Fiedorczuk / Leonid A Sazanov /   Abstract: Mitochondrial electron transport chain complexes are organized into supercomplexes responsible for carrying out cellular respiration. Here we present three architectures of mammalian (ovine) ...Mitochondrial electron transport chain complexes are organized into supercomplexes responsible for carrying out cellular respiration. Here we present three architectures of mammalian (ovine) supercomplexes determined by cryo-electron microscopy. We identify two distinct arrangements of supercomplex CICIIICIV (the respirasome)-a major 'tight' form and a minor 'loose' form (resolved at the resolution of 5.8 Å and 6.7 Å, respectively), which may represent different stages in supercomplex assembly or disassembly. We have also determined an architecture of supercomplex CICIII at 7.8 Å resolution. All observed density can be attributed to the known 80 subunits of the individual complexes, including 132 transmembrane helices. The individual complexes form tight interactions that vary between the architectures, with complex IV subunit COX7a switching contact from complex III to complex I. The arrangement of active sites within the supercomplex may help control reactive oxygen species production. To our knowledge, these are the first complete architectures of the dominant, physiologically relevant state of the electron transport chain. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8130.map.gz emd_8130.map.gz | 436.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8130-v30.xml emd-8130-v30.xml emd-8130.xml emd-8130.xml | 31.6 KB 31.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8130_fsc.xml emd_8130_fsc.xml | 17.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_8130.png emd_8130.png | 126.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8130 http://ftp.pdbj.org/pub/emdb/structures/EMD-8130 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8130 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8130 | HTTPS FTP |
-Related structure data
| Related structure data |  5j4zMC  8128C  8129C  5j7yC  5j8kC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8130.map.gz / Format: CCP4 / Size: 465.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8130.map.gz / Format: CCP4 / Size: 465.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.72 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ovine respiratory supercomplex (I-III2-IV), conformation tight
| Entire | Name: Ovine respiratory supercomplex (I-III2-IV), conformation tight |
|---|---|
| Components |
|
-Supramolecule #1: Ovine respiratory supercomplex (I-III2-IV), conformation tight
| Supramolecule | Name: Ovine respiratory supercomplex (I-III2-IV), conformation tight type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#68 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 1.71 MDa |
-Supramolecule #2: Respiratory complex IV
| Supramolecule | Name: Respiratory complex IV / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#13 / Details: Monomer |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 220 KDa |
-Supramolecule #3: Respiratory complex III
| Supramolecule | Name: Respiratory complex III / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #14-#24 / Details: Dimer |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 490 KDa |
-Supramolecule #4: Respiratory complex I
| Supramolecule | Name: Respiratory complex I / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #25-#68 / Details: Monomer |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.0 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.7 Component:
| ||||||||||||
| Grid | Model: Quantifoil R0.6/1 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Blotted 32 seconds. | ||||||||||||
| Details | Single particles dispersed in digitonin |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Sampling interval: 14.0 µm / Digitization - Frames/image: 1-32 / Number grids imaged: 3 / Number real images: 1608 / Average exposure time: 4.0 sec. / Average electron dose: 34.0 e/Å2 Details: Images were collected in movie mode with 17 frames per second. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 81395 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 47000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: BACKBONE TRACE |
|---|---|
| Output model |  PDB-5j4z: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)