+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8070 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Yeast V-ATPase average of densities, a subunit segment | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | V-ATPase / Vo region / membrane protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to alkaline pH / protein localization to vacuolar membrane / Insulin receptor recycling / Transferrin endocytosis and recycling / polyphosphate metabolic process / ROS and RNS production in phagocytes / Amino acids regulate mTORC1 / vacuolar proton-transporting V-type ATPase, V0 domain / fungal-type vacuole / vacuolar proton-transporting V-type ATPase complex ...cellular response to alkaline pH / protein localization to vacuolar membrane / Insulin receptor recycling / Transferrin endocytosis and recycling / polyphosphate metabolic process / ROS and RNS production in phagocytes / Amino acids regulate mTORC1 / vacuolar proton-transporting V-type ATPase, V0 domain / fungal-type vacuole / vacuolar proton-transporting V-type ATPase complex / cellular hyperosmotic response / vacuolar acidification / fungal-type vacuole membrane / phosphatidylinositol-3,5-bisphosphate binding / proton-transporting ATPase activity, rotational mechanism / Neutrophil degranulation / proton transmembrane transport / ATPase binding / protein-containing complex assembly / membrane raft Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.0 Å | |||||||||
 Authors Authors | Schep DG / Zhao J | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2016 Journal: Proc Natl Acad Sci U S A / Year: 2016Title: Models for the a subunits of the Thermus thermophilus V/A-ATPase and Saccharomyces cerevisiae V-ATPase enzymes by cryo-EM and evolutionary covariance. Authors: Daniel G Schep / Jianhua Zhao / John L Rubinstein /  Abstract: Rotary ATPases couple ATP synthesis or hydrolysis to proton translocation across a membrane. However, understanding proton translocation has been hampered by a lack of structural information for the ...Rotary ATPases couple ATP synthesis or hydrolysis to proton translocation across a membrane. However, understanding proton translocation has been hampered by a lack of structural information for the membrane-embedded a subunit. The V/A-ATPase from the eubacterium Thermus thermophilus is similar in structure to the eukaryotic V-ATPase but has a simpler subunit composition and functions in vivo to synthesize ATP rather than pump protons. We determined the T. thermophilus V/A-ATPase structure by cryo-EM at 6.4 Å resolution. Evolutionary covariance analysis allowed tracing of the a subunit sequence within the map, providing a complete model of the rotary ATPase. Comparing the membrane-embedded regions of the T. thermophilus V/A-ATPase and eukaryotic V-ATPase from Saccharomyces cerevisiae allowed identification of the α-helices that belong to the a subunit and revealed the existence of previously unknown subunits in the eukaryotic enzyme. Subsequent evolutionary covariance analysis enabled construction of a model of the a subunit in the S. cerevisae V-ATPase that explains numerous biochemical studies of that enzyme. Comparing the two a subunit structures determined here with a structure of the distantly related a subunit from the bovine F-type ATP synthase revealed a conserved pattern of residues, suggesting a common mechanism for proton transport in all rotary ATPases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8070.map.gz emd_8070.map.gz | 112.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8070-v30.xml emd-8070-v30.xml emd-8070.xml emd-8070.xml | 12.9 KB 12.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8070.png emd_8070.png | 49.9 KB | ||
| Filedesc metadata |  emd-8070.cif.gz emd-8070.cif.gz | 5.4 KB | ||
| Others |  emd_8070_additional.map.gz emd_8070_additional.map.gz | 603.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8070 http://ftp.pdbj.org/pub/emdb/structures/EMD-8070 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8070 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8070 | HTTPS FTP |
-Related structure data
| Related structure data |  5i1mMC  8016C  8017C  5garC  5gasC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8070.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8070.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.45 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
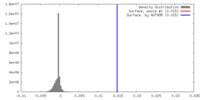
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: None
| File | emd_8070_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||
| Projections & Slices |
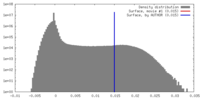
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : V-ATPase
| Entire | Name: V-ATPase |
|---|---|
| Components |
|
-Supramolecule #1: V-ATPase
| Supramolecule | Name: V-ATPase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Six maps of the V-ATPase, averaged with the Vo regions aligned |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: V-ATPase a subunit
| Supramolecule | Name: V-ATPase a subunit / type: complex / ID: 2 / Parent: 1 / Macromolecule list: all Details: Segment of averaged densities from six maps of the V-ATPase, aligned to the a subunit. This does not contain the c ring or micelle, but has some densities that are not assigned to the a ...Details: Segment of averaged densities from six maps of the V-ATPase, aligned to the a subunit. This does not contain the c ring or micelle, but has some densities that are not assigned to the a subunit and likely represent unknown subunits. |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: V-type proton ATPase subunit a, vacuolar isoform
| Macromolecule | Name: V-type proton ATPase subunit a, vacuolar isoform / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 51.548867 KDa |
| Sequence | String: TNKFTAGFQS ICDCYGIAQY REINAGLPTI VTFPFMFAIM FGDMGHGFLM TLAALSLVLN EKKINKMKRG EIFDMAFTGR YIILLMGVF SMYTGFLYND IFSKTMTIFK SGWKWPDHWK KGESITATSV GTYPIGLDWA WHGTENALLF SNSYKMKLSI L MGFIHMTY ...String: TNKFTAGFQS ICDCYGIAQY REINAGLPTI VTFPFMFAIM FGDMGHGFLM TLAALSLVLN EKKINKMKRG EIFDMAFTGR YIILLMGVF SMYTGFLYND IFSKTMTIFK SGWKWPDHWK KGESITATSV GTYPIGLDWA WHGTENALLF SNSYKMKLSI L MGFIHMTY SYFFSLANHL YFNSMIDIIG NFIPGLLFMQ GIFGYLSVCI VYKWAVDWVK DGKPAPGLLN MLINMFLSPG TI DDELYPH QAKVQVFLLL MALVCIPWLL LVKPLHFKFT HKKKSHEPLP STEADASSED LEAQQLISAM DADDAEEEEV GSG SHGEDF GDIMIHQVIH TIEFCLNCVS HTASYLRLWA LSLAHAQLSS VLWTMTIQIA FGFRGFVGVF MTVALFAMWF ALTC AVLVL MEGTSAMLHS LRLHWVESMS KFFVGEGLPY EPFAFEYKDM EVAVASASSS ASS UniProtKB: V-type proton ATPase subunit a, vacuolar isoform |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 35.7 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 7.0 Å / Resolution method: OTHER Details: Resolution is approximated from the resolutions of the aligned and averaged maps that yielded the density. Number images used: 269377 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)