[English] 日本語
 Yorodumi
Yorodumi- EMDB-7960: Open state GluA2 in complex with STZ and blocked by IEM-1460, aft... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7960 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Open state GluA2 in complex with STZ and blocked by IEM-1460, after micelle signal subtraction | ||||||||||||
 Map data Map data | TMD-directed refinement after micelle signal subtraction | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Ion channel / TRANSPORT PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationLGI-ADAM interactions / Presynaptic depolarization and calcium channel opening / eye blink reflex / positive regulation of protein localization to basolateral plasma membrane / cerebellar mossy fiber / postsynaptic neurotransmitter receptor diffusion trapping / regulation of AMPA receptor activity / channel regulator activity / Trafficking of AMPA receptors / membrane hyperpolarization ...LGI-ADAM interactions / Presynaptic depolarization and calcium channel opening / eye blink reflex / positive regulation of protein localization to basolateral plasma membrane / cerebellar mossy fiber / postsynaptic neurotransmitter receptor diffusion trapping / regulation of AMPA receptor activity / channel regulator activity / Trafficking of AMPA receptors / membrane hyperpolarization / protein targeting to membrane / voltage-gated calcium channel complex / spine synapse / dendritic spine neck / dendritic spine head / cellular response to amine stimulus / neurotransmitter receptor localization to postsynaptic specialization membrane / neuromuscular junction development / perisynaptic space / Activation of AMPA receptors / ligand-gated monoatomic cation channel activity / AMPA glutamate receptor activity / transmission of nerve impulse / response to lithium ion / Trafficking of GluR2-containing AMPA receptors / kainate selective glutamate receptor activity / cellular response to glycine / AMPA glutamate receptor complex / extracellularly glutamate-gated ion channel activity / immunoglobulin binding / asymmetric synapse / ionotropic glutamate receptor complex / conditioned place preference / regulation of receptor recycling / membrane depolarization / glutamate receptor binding / Unblocking of NMDA receptors, glutamate binding and activation / positive regulation of synaptic transmission / positive regulation of synaptic transmission, glutamatergic / regulation of synaptic transmission, glutamatergic / voltage-gated calcium channel activity / response to fungicide / cytoskeletal protein binding / glutamate-gated receptor activity / regulation of long-term synaptic depression / extracellular ligand-gated monoatomic ion channel activity / cellular response to brain-derived neurotrophic factor stimulus / glutamate-gated calcium ion channel activity / presynaptic active zone membrane / somatodendritic compartment / dendrite membrane / ionotropic glutamate receptor binding / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / ionotropic glutamate receptor signaling pathway / dendrite cytoplasm / synaptic membrane / hippocampal mossy fiber to CA3 synapse / dendritic shaft / SNARE binding / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / synaptic transmission, glutamatergic / protein tetramerization / PDZ domain binding / establishment of protein localization / response to calcium ion / postsynaptic density membrane / cerebral cortex development / modulation of chemical synaptic transmission / receptor internalization / Schaffer collateral - CA1 synapse / terminal bouton / synaptic vesicle / endocytic vesicle membrane / synaptic vesicle membrane / presynapse / signaling receptor activity / amyloid-beta binding / presynaptic membrane / growth cone / scaffold protein binding / perikaryon / chemical synaptic transmission / dendritic spine / postsynaptic membrane / neuron projection / postsynaptic density / axon / external side of plasma membrane / neuronal cell body / dendrite / synapse / protein kinase binding / protein-containing complex binding / glutamatergic synapse / cell surface / endoplasmic reticulum / protein-containing complex / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | ||||||||||||
 Authors Authors | Twomey EC / Yelshanskaya MV | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Neuron / Year: 2018 Journal: Neuron / Year: 2018Title: Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors. Authors: Edward C Twomey / Maria V Yelshanskaya / Alexander A Vassilevski / Alexander I Sobolevsky /   Abstract: AMPA receptors mediate fast excitatory neurotransmission and are critical for CNS development and function. Calcium-permeable subsets of AMPA receptors are strongly implicated in acute and chronic ...AMPA receptors mediate fast excitatory neurotransmission and are critical for CNS development and function. Calcium-permeable subsets of AMPA receptors are strongly implicated in acute and chronic neurological disorders. However, despite the clinical importance, the therapeutic landscape for specifically targeting them, and not the calcium-impermeable AMPA receptors, remains largely undeveloped. To address this problem, we used cryo-electron microscopy and electrophysiology to investigate the mechanisms by which small-molecule blockers selectively inhibit ion channel conductance in calcium-permeable AMPA receptors. We determined the structures of calcium-permeable GluA2 AMPA receptor complexes with the auxiliary subunit stargazin bound to channel blockers, including the orb weaver spider toxin AgTx-636, the spider toxin analog NASPM, and the adamantane derivative IEM-1460. Our structures provide insights into the architecture of the blocker binding site and the mechanism of trapping, which are critical for development of small molecules that specifically target calcium-permeable AMPA receptors. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7960.map.gz emd_7960.map.gz | 9.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7960-v30.xml emd-7960-v30.xml emd-7960.xml emd-7960.xml | 15.1 KB 15.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7960.png emd_7960.png | 274.5 KB | ||
| Filedesc metadata |  emd-7960.cif.gz emd-7960.cif.gz | 6.4 KB | ||
| Others |  emd_7960_additional.map.gz emd_7960_additional.map.gz | 6.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7960 http://ftp.pdbj.org/pub/emdb/structures/EMD-7960 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7960 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7960 | HTTPS FTP |
-Validation report
| Summary document |  emd_7960_validation.pdf.gz emd_7960_validation.pdf.gz | 386.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7960_full_validation.pdf.gz emd_7960_full_validation.pdf.gz | 385.6 KB | Display | |
| Data in XML |  emd_7960_validation.xml.gz emd_7960_validation.xml.gz | 6.4 KB | Display | |
| Data in CIF |  emd_7960_validation.cif.gz emd_7960_validation.cif.gz | 7.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7960 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7960 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7960 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7960 | HTTPS FTP |
-Related structure data
| Related structure data |  6dm0MC  7959C  7961C  7962C  6dlzC  6dm1C  6o9gC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7960.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7960.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TMD-directed refinement after micelle signal subtraction | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Refinement of full map after micelle signal subtraction
| File | emd_7960_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refinement of full map after micelle signal subtraction | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Open state GluA2 in complex with STZ and blocked by IEM-1460, aft...
| Entire | Name: Open state GluA2 in complex with STZ and blocked by IEM-1460, after micelle signal subtraction |
|---|---|
| Components |
|
-Supramolecule #1: Open state GluA2 in complex with STZ and blocked by IEM-1460, aft...
| Supramolecule | Name: Open state GluA2 in complex with STZ and blocked by IEM-1460, after micelle signal subtraction type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: The full-map (non-TMD-directed) is included as a supplemental file |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit
| Macromolecule | Name: Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 115.178531 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NSIQIGGLFP RGADQEYSAF RVGMVQFSTS EFRLTPHIDN LEVANSFAVT NAFCSQFSRG VYAIFGFYDK KSVNTITSFC GTLHVSFIT PSFPTDGTHP FVIQMRPDLK GALLSLIEYY QWDKFAYLYD SDRGLSTLQA VLDSAAEKKW QVTAINVGNI N NDKKDETY ...String: NSIQIGGLFP RGADQEYSAF RVGMVQFSTS EFRLTPHIDN LEVANSFAVT NAFCSQFSRG VYAIFGFYDK KSVNTITSFC GTLHVSFIT PSFPTDGTHP FVIQMRPDLK GALLSLIEYY QWDKFAYLYD SDRGLSTLQA VLDSAAEKKW QVTAINVGNI N NDKKDETY RSLFQDLELK KERRVILDCE RDKVNDIVDQ VITIGKHVKG YHYIIANLGF TDGDLLKIQF GGAEVSGFQI VD YDDSLVS KFIERWSTLE EKEYPGAHTA TIKYTSALTY DAVQVMTEAF RNLRKQRIEI SRRGNAGDCL ANPAVPWGQG VEI ERALKQ VQVEGLSGNI KFDQNGKRIN YTINIMELKT NGPRKIGYWS EVDKMVLTED DTSGLEQKTV VVTTILESPY VMMK KNHEM LEGNERYEGY CVDLAAEIAK HCGFKYKLTI VGDGKYGARD ADTKIWNGMV GELVYGKADI AIAPLTITLV REEVI DFSK PFMSLGISIM IKKPQKSKPG VFSFLDPLAY EIWMCIVFAY IGVSVVLFLV SRFSPYEWHT EEFEDGRETQ SSESTN EFG IFNSLWFSLG AFMQQGCDIS PRSLSGRIVG GVWWFFTLII ISSYTANLAA FLTVERMVSP IESAEDLSKQ TEIAYGT LD SGSTKEFFRR SKIAVFDKMW TYMRSAEPSV FVRTTAEGVA RVRKSKGKYA YLLESTMNEY IEQRKPCDTM KVGGNLDS K GYGIATPKGS SLGTPVNLAV LKLSEQGVLD KLKNKWWYDK GECGAKDSGS KEKTSALSLS NVAGVFYILV GGLGLAMLV ALIEFCYKSR AEAKRMKGTG LFDRGVQMLL TTVGAFAAFS LMTIAVGTDY WLYSRGVCKT KSVSEDETSK KNEEVMTHSG LWRTCCLEG NFKGLCKQID HFPEDADYEA DTAEYFLRAV RASSIFPILS VILLFMGGLC IAASEFYKTR HNIILSAGIF F VSAGLSNI IGIIVYISAN AGDPSKSDSK KNSYSYGWSF YFGALSFIIA EMVGVLAVHM FIDRHKQLTG GAE UniProtKB: Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit |
-Macromolecule #2: GLUTAMIC ACID
| Macromolecule | Name: GLUTAMIC ACID / type: ligand / ID: 2 / Number of copies: 4 / Formula: GLU |
|---|---|
| Molecular weight | Theoretical: 147.129 Da |
| Chemical component information |  ChemComp-GLU: |
-Macromolecule #3: CYCLOTHIAZIDE
| Macromolecule | Name: CYCLOTHIAZIDE / type: ligand / ID: 3 / Number of copies: 4 / Formula: CYZ |
|---|---|
| Molecular weight | Theoretical: 389.878 Da |
| Chemical component information | 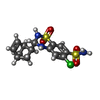 ChemComp-CYZ: |
-Macromolecule #4: N,N,N-trimethyl-5-({[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]decan-1-yl]...
| Macromolecule | Name: N,N,N-trimethyl-5-({[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]decan-1-yl]methyl}amino)pentan-1-aminium type: ligand / ID: 4 / Number of copies: 1 / Formula: GZD |
|---|---|
| Molecular weight | Theoretical: 293.51 Da |
| Chemical component information | 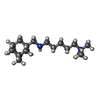 ChemComp-GZD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
| Details | Open state GluA2 in complex with STZ and blocked by IEM-1460, after micelle signal subtraction |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average exposure time: 8.0 sec. / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: Details: Activated GluA2 complex bound to glutamate, cyclothiazide, and STZ in digitonin |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 4.4 Å / Resolution method: FSC 0.143 CUT-OFF Details: Full map resolution (supplemental file) is 4.8 angstrom. Number images used: 30849 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-6dm0: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)