+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7612 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
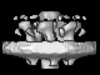
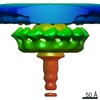
| Title | Cytoplasmic ATPase in a Type IV Secretion Machine | |||||||||
 Map data Map data | Cytoplasmic ATPase complex | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 33.0 Å | |||||||||
 Authors Authors | Liu J / Hu B | |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2018 Journal: Nat Microbiol / Year: 2018Title: A unique cytoplasmic ATPase complex defines the Legionella pneumophila type IV secretion channel. Authors: David Chetrit / Bo Hu / Peter J Christie / Craig R Roy / Jun Liu /  Abstract: Type IV secretion systems (T4SSs) are complex machines used by bacteria to deliver protein and DNA complexes into target host cells. Conserved ATPases are essential for T4SS function, but how they ...Type IV secretion systems (T4SSs) are complex machines used by bacteria to deliver protein and DNA complexes into target host cells. Conserved ATPases are essential for T4SS function, but how they coordinate their activities to promote substrate transfer remains poorly understood. Here, we show that the DotB ATPase associates with the Dot-Icm T4SS at the Legionella cell pole through interactions with the DotO ATPase. The structure of the Dot-Icm apparatus was solved in situ by cryo-electron tomography at 3.5 nm resolution and the cytoplasmic complex was solved at 3.0 nm resolution. These structures revealed a cell envelope-spanning channel that connects to the cytoplasmic complex. Further analysis revealed a hexameric assembly of DotO dimers associated with the inner membrane complex, and a DotB hexamer associated with the base of this cytoplasmic complex. The assembly of a DotB-DotO energy complex creates a cytoplasmic channel that directs the translocation of substrates through the T4SS. These data define distinct stages in Dot-Icm machine biogenesis, advance our understanding of channel activation, and identify an envelope-spanning T4SS channel. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7612.map.gz emd_7612.map.gz | 1.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7612-v30.xml emd-7612-v30.xml emd-7612.xml emd-7612.xml | 8 KB 8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7612.png emd_7612.png | 94.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7612 http://ftp.pdbj.org/pub/emdb/structures/EMD-7612 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7612 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7612 | HTTPS FTP |
-Validation report
| Summary document |  emd_7612_validation.pdf.gz emd_7612_validation.pdf.gz | 78.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7612_full_validation.pdf.gz emd_7612_full_validation.pdf.gz | 77.2 KB | Display | |
| Data in XML |  emd_7612_validation.xml.gz emd_7612_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7612 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7612 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7612 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7612 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7612.map.gz / Format: CCP4 / Size: 1.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7612.map.gz / Format: CCP4 / Size: 1.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cytoplasmic ATPase complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
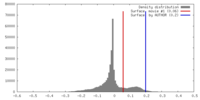
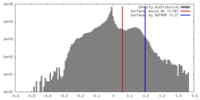
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Type IV secretion machine
| Entire | Name: Type IV secretion machine |
|---|---|
| Components |
|
-Supramolecule #1: Type IV secretion machine
| Supramolecule | Name: Type IV secretion machine / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average exposure time: 1.0 sec. / Average electron dose: 1.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 33.0 Å / Resolution method: FSC 0.5 CUT-OFF / Number subtomograms used: 5000 |
|---|---|
| Extraction | Number tomograms: 1000 / Number images used: 5000 / Method: manually |
| CTF correction | Software - Name:  IMOD (ver. 4.9.6) IMOD (ver. 4.9.6) |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)