+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7543 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Zika virus at a resolution of 3.1 Angstrom | |||||||||
 Map data Map data | ZIKV map at resolution of 3.1 Angstrom | |||||||||
 Sample Sample | Zika virus != Zika virus ZIKV/H. sapiens/FrenchPolynesia/10087PF/2013 Zika virus
| |||||||||
 Keywords Keywords | Zika / flavivirus / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationflavivirin / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of host TYK2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / negative regulation of innate immune response / viral capsid / double-stranded RNA binding / nucleoside-triphosphate phosphatase / 4 iron, 4 sulfur cluster binding / mRNA (guanine-N7)-methyltransferase ...flavivirin / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of host TYK2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / negative regulation of innate immune response / viral capsid / double-stranded RNA binding / nucleoside-triphosphate phosphatase / 4 iron, 4 sulfur cluster binding / mRNA (guanine-N7)-methyltransferase / methyltransferase cap1 / molecular adaptor activity / clathrin-dependent endocytosis of virus by host cell / methyltransferase cap1 activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / RNA helicase activity / protein dimerization activity / host cell perinuclear region of cytoplasm / host cell endoplasmic reticulum membrane / RNA helicase / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / symbiont-mediated activation of host autophagy / serine-type endopeptidase activity / RNA-directed RNA polymerase / viral RNA genome replication / RNA-directed RNA polymerase activity / fusion of virus membrane with host endosome membrane / lipid binding / viral envelope / centrosome / GTP binding / virion attachment to host cell / host cell nucleus / virion membrane / structural molecule activity / ATP hydrolysis activity / proteolysis / extracellular region / ATP binding / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Zika virus ZIKV/H. sapiens/FrenchPolynesia/10087PF/2013 Zika virus ZIKV/H. sapiens/FrenchPolynesia/10087PF/2013 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Sevvana M / Long F | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2016 Journal: Science / Year: 2016Title: The 3.8 Å resolution cryo-EM structure of Zika virus. Authors: Devika Sirohi / Zhenguo Chen / Lei Sun / Thomas Klose / Theodore C Pierson / Michael G Rossmann / Richard J Kuhn /  Abstract: The recent rapid spread of Zika virus and its unexpected linkage to birth defects and an autoimmune neurological syndrome have generated worldwide concern. Zika virus is a flavivirus like the dengue, ...The recent rapid spread of Zika virus and its unexpected linkage to birth defects and an autoimmune neurological syndrome have generated worldwide concern. Zika virus is a flavivirus like the dengue, yellow fever, and West Nile viruses. We present the 3.8 angstrom resolution structure of mature Zika virus, determined by cryo-electron microscopy (cryo-EM). The structure of Zika virus is similar to other known flavivirus structures, except for the ~10 amino acids that surround the Asn(154) glycosylation site in each of the 180 envelope glycoproteins that make up the icosahedral shell. The carbohydrate moiety associated with this residue, which is recognizable in the cryo-EM electron density, may function as an attachment site of the virus to host cells. This region varies not only among Zika virus strains but also in other flaviviruses, which suggests that differences in this region may influence virus transmission and disease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7543.map.gz emd_7543.map.gz | 3.8 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7543-v30.xml emd-7543-v30.xml emd-7543.xml emd-7543.xml | 24.3 KB 24.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7543_fsc_1.xml emd_7543_fsc_1.xml emd_7543_fsc_2.xml emd_7543_fsc_2.xml | 33.9 KB 33.9 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_7543.png emd_7543.png | 218.2 KB | ||
| Filedesc metadata |  emd-7543.cif.gz emd-7543.cif.gz | 6.4 KB | ||
| Others |  emd_7543_additional_1.map.gz emd_7543_additional_1.map.gz emd_7543_additional_2.map.gz emd_7543_additional_2.map.gz emd_7543_half_map_1.map.gz emd_7543_half_map_1.map.gz emd_7543_half_map_2.map.gz emd_7543_half_map_2.map.gz | 543.9 MB 543.9 MB 239.3 MB 240.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7543 http://ftp.pdbj.org/pub/emdb/structures/EMD-7543 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7543 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7543 | HTTPS FTP |
-Validation report
| Summary document |  emd_7543_validation.pdf.gz emd_7543_validation.pdf.gz | 811.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7543_full_validation.pdf.gz emd_7543_full_validation.pdf.gz | 811 KB | Display | |
| Data in XML |  emd_7543_validation.xml.gz emd_7543_validation.xml.gz | 31.6 KB | Display | |
| Data in CIF |  emd_7543_validation.cif.gz emd_7543_validation.cif.gz | 36.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7543 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7543 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7543 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7543 | HTTPS FTP |
-Related structure data
| Related structure data |  6co8MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7543.map.gz / Format: CCP4 / Size: 4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7543.map.gz / Format: CCP4 / Size: 4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ZIKV map at resolution of 3.1 Angstrom | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
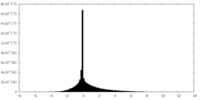
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: ZIKV, unmasked half map - even
| File | emd_7543_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ZIKV, unmasked half map - even | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: ZIKV, unmasked half map - odd
| File | emd_7543_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ZIKV, unmasked half map - odd | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: ZIKV, masked half map - even
| File | emd_7543_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ZIKV, masked half map - even | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: ZIKV, masked half map - odd
| File | emd_7543_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ZIKV, masked half map - odd | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Zika virus
| Entire | Name:   Zika virus Zika virus |
|---|---|
| Components |
|
-Supramolecule #1: Zika virus ZIKV/H. sapiens/FrenchPolynesia/10087PF/2013
| Supramolecule | Name: Zika virus ZIKV/H. sapiens/FrenchPolynesia/10087PF/2013 type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2043570 Sci species name: Zika virus ZIKV/H. sapiens/FrenchPolynesia/10087PF/2013 Sci species strain: ZIKV/Human/French Polynesia/10087PF/2013 Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Virus shell | Shell ID: 1 / Diameter: 500.0 Å / T number (triangulation number): 3 |
-Macromolecule #1: E protein
| Macromolecule | Name: E protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Zika virus ZIKV/H. sapiens/FrenchPolynesia/10087PF/2013 Zika virus ZIKV/H. sapiens/FrenchPolynesia/10087PF/2013 |
| Molecular weight | Theoretical: 54.444051 KDa |
| Sequence | String: IRCIGVSNRD FVEGMSGGTW VDVVLEHGGC VTVMAQDKPT VDIELVTTTV SNMAEVRSYC YEASISDMAS DSRCPTQGEA YLDKQSDTQ YVCKRTLVDR GWGNGCGLFG KGSLVTCAKF ACSKKMTGKS IQPENLEYRI MLSVHGSQHS GMIVNDTGHE T DENRAKVE ...String: IRCIGVSNRD FVEGMSGGTW VDVVLEHGGC VTVMAQDKPT VDIELVTTTV SNMAEVRSYC YEASISDMAS DSRCPTQGEA YLDKQSDTQ YVCKRTLVDR GWGNGCGLFG KGSLVTCAKF ACSKKMTGKS IQPENLEYRI MLSVHGSQHS GMIVNDTGHE T DENRAKVE ITPNSPRAEA TLGGFGSLGL DCEPRTGLDF SDLYYLTMNN KHWLVHKEWF HDIPLPWHAG ADTGTPHWNN KE ALVEFKD AHAKRQTVVV LGSQEGAVHT ALAGALEAEM DGAKGRLSSG HLKCRLKMDK LRLKGVSYSL CTAAFTFTKI PAE TLHGTV TVEVQYAGTD GPCKVPAQMA VDMQTLTPVG RLITANPVIT ESTENSKMML ELDPPFGDSY IVIGVGEKKI THHW HRSGS TIGKAFEATV RGAKRMAVLG DTAWDFGSVG GALNSLGKGI HQIFGAAFKS LFGGMSWFSQ ILIGTLLMWL GLNTK NGSI SLMCLALGGV LIFLSTAVSA UniProtKB: Genome polyprotein |
-Macromolecule #2: M protein
| Macromolecule | Name: M protein / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Zika virus ZIKV/H. sapiens/FrenchPolynesia/10087PF/2013 Zika virus ZIKV/H. sapiens/FrenchPolynesia/10087PF/2013 |
| Molecular weight | Theoretical: 8.496883 KDa |
| Sequence | String: AVTLPSHSTR KLQTRSQTWL ESREYTKHLI RVENWIFRNP GFALAAAAIA WLLGSSTSQK VIYLVMILLI APAYS UniProtKB: Genome polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 12 mM Tris-HCl, pH 8, 120 mM NaCl, 1 mM EDTA |
| Grid | Mesh: 400 / Support film - Material: CARBON / Support film - topology: LACEY |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 100.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 7420 pixel / Digitization - Dimensions - Height: 7676 pixel / Number grids imaged: 1 / Number real images: 2085 / Average exposure time: 11.0 sec. / Average electron dose: 33.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 30864 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 18000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Real space followed by reciprocal space |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-6co8: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)