[English] 日本語
 Yorodumi
Yorodumi- EMDB-7513: Subtomogram average of mature CHIKV virion budded from human cells -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7513 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram average of mature CHIKV virion budded from human cells | |||||||||
 Map data Map data | Mature CHIKV virion budded from human cells | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Chikungunya virus Chikungunya virus | |||||||||
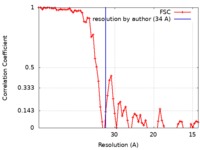
| Method | subtomogram averaging / cryo EM / Resolution: 34.0 Å | |||||||||
 Authors Authors | Galaz-Montoya JG / Jin J / Sherman MB | |||||||||
 Citation Citation |  Journal: Cell Host Microbe / Year: 2018 Journal: Cell Host Microbe / Year: 2018Title: Neutralizing Antibodies Inhibit Chikungunya Virus Budding at the Plasma Membrane. Authors: Jing Jin / Jesús G Galaz-Montoya / Michael B Sherman / Stella Y Sun / Cynthia S Goldsmith / Eileen T O'Toole / Larry Ackerman / Lars-Anders Carlson / Scott C Weaver / Wah Chiu / Graham Simmons /   Abstract: Neutralizing antibodies (NAbs) are traditionally thought to inhibit virus infection by preventing virion entry into target cells. In addition, antibodies can engage Fc receptors (FcRs) on immune ...Neutralizing antibodies (NAbs) are traditionally thought to inhibit virus infection by preventing virion entry into target cells. In addition, antibodies can engage Fc receptors (FcRs) on immune cells to activate antiviral responses. We describe a mechanism by which NAbs inhibit chikungunya virus (CHIKV), the most common alphavirus infecting humans, by preventing virus budding from infected human cells and activating IgG-specific Fcγ receptors. NAbs bind to CHIKV glycoproteins on the infected cell surface and induce glycoprotein coalescence, preventing budding of nascent virions and leaving structurally heterogeneous nucleocapsids arrested in the cytosol. Furthermore, NAbs induce clustering of CHIKV replication spherules at sites of budding blockage. Functionally, these densely packed glycoprotein-NAb complexes on infected cells activate Fcγ receptors, inducing a strong, antibody-dependent, cell-mediated cytotoxicity response from immune effector cells. Our findings describe a triply functional antiviral pathway for NAbs that might be broadly applicable across virus-host systems, suggesting avenues for therapeutic innovation through antibody design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7513.map.gz emd_7513.map.gz | 883.9 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7513-v30.xml emd-7513-v30.xml emd-7513.xml emd-7513.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7513_fsc.xml emd_7513_fsc.xml | 6.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_7513.png emd_7513.png | 175.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7513 http://ftp.pdbj.org/pub/emdb/structures/EMD-7513 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7513 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7513 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7513.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7513.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mature CHIKV virion budded from human cells | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 7.16 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Chikungunya virus
| Entire | Name:   Chikungunya virus Chikungunya virus |
|---|---|
| Components |
|
-Supramolecule #1: Chikungunya virus
| Supramolecule | Name: Chikungunya virus / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 37124 / Sci species name: Chikungunya virus / Sci species strain: vaccine strain 181/clone 25 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) / Strain: vaccine strain 181/clone 25 Homo sapiens (human) / Strain: vaccine strain 181/clone 25 |
| Virus shell | Shell ID: 1 / Diameter: 698.0 Å / T number (triangulation number): 4 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Details | CHIKV vaccine strain 181/clone 25 infected U2OS cells. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2200FS |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Average exposure time: 0.6 sec. / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)