+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1692 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM reconstruction of Phage Gifsy-2 darker procapsid | |||||||||
 Map data Map data | Gifsy-2 darker procapsid | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bacteriophage / Gifsy-2 / capsid | |||||||||
| Biological species |  Phage Gifsy-2 (virus) Phage Gifsy-2 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 25.6 Å | |||||||||
 Authors Authors | Effantin G / Figueroa-Bossi N / Schoehn G / Bossi L / Conway JF | |||||||||
 Citation Citation |  Journal: Virology / Year: 2010 Journal: Virology / Year: 2010Title: The tripartite capsid gene of Salmonella phage Gifsy-2 yields a capsid assembly pathway engaging features from HK97 and lambda. Authors: Grégory Effantin / Nara Figueroa-Bossi / Guy Schoehn / Lionello Bossi / James F Conway /  Abstract: Phage Gifsy-2, a lambdoid phage infecting Salmonella, has an unusually large composite gene coding for its major capsid protein (mcp) at the C-terminal end, a ClpP-like protease at the N-terminus, ...Phage Gifsy-2, a lambdoid phage infecting Salmonella, has an unusually large composite gene coding for its major capsid protein (mcp) at the C-terminal end, a ClpP-like protease at the N-terminus, and a approximately 200 residue central domain of unknown function but which may have a scaffolding role. This combination of functions on a single coding region is more extensive than those observed in other phages such as HK97 (scaffold-capsid fusion) and lambda (protease-scaffold fusion). To study the structural phenotype of the unique Gifsy-2 capsid gene, we have purified Gifsy-2 particles and visualized capsids and procapsids by cryoelectron microscopy, determining structures to resolutions up to 12A. The capsids have lambdoid T=7 geometry and are well modeled with the atomic structures of HK97 mcp and phage lambda gpD decoration protein. Thus, the unique Gifsy-2 capsid protein gene yields a capsid maturation pathway engaging features from both phages HK97 and lambda. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1692.map.gz emd_1692.map.gz | 34 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1692-v30.xml emd-1692-v30.xml emd-1692.xml emd-1692.xml | 8.1 KB 8.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd-1692.png emd-1692.png | 512.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1692 http://ftp.pdbj.org/pub/emdb/structures/EMD-1692 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1692 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1692 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1692.map.gz / Format: CCP4 / Size: 95.1 MB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) Download / File: emd_1692.map.gz / Format: CCP4 / Size: 95.1 MB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Gifsy-2 darker procapsid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.842 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Bacteriophage Gifsy-2 procapsid purified from Salmonella Typhimurium
| Entire | Name: Bacteriophage Gifsy-2 procapsid purified from Salmonella Typhimurium |
|---|---|
| Components |
|
-Supramolecule #1000: Bacteriophage Gifsy-2 procapsid purified from Salmonella Typhimurium
| Supramolecule | Name: Bacteriophage Gifsy-2 procapsid purified from Salmonella Typhimurium type: sample / ID: 1000 / Oligomeric state: 1 / Number unique components: 1 |
|---|
-Supramolecule #1: Phage Gifsy-2
| Supramolecule | Name: Phage Gifsy-2 / type: virus / ID: 1 / Name.synonym: Gifsy-2 / NCBI-ID: 129862 / Sci species name: Phage Gifsy-2 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No / Syn species name: Gifsy-2 |
|---|---|
| Host (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria)synonym: BACTERIA(EUBACTERIA) |
| Virus shell | Shell ID: 1 / Diameter: 570 Å / T number (triangulation number): 7 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 10 mM Tris-HCl pH7.5, 10 mM MgSO4 |
|---|---|
| Grid | Details: 300 mesh copper grid |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: Home made manual plunger |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200T |
|---|---|
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 1.84 µm / Number real images: 51 |
| Electron beam | Acceleration voltage: 200 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 38000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 25.6 Å / Resolution method: FSC 0.33 CUT-OFF / Software - Name: pft - em3dr / Number images used: 56 |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera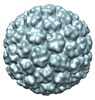













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















