+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7473 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subparticle refinement of HSV-1 capsid vertex region. | |||||||||||||||||||||
 Map data Map data | Subparticle refinement of HSV-1 capsid vertex region with associated tegument proteins. | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationviral genome packaging / T=16 icosahedral viral capsid / deNEDDylase activity / viral tegument / viral capsid assembly / viral DNA genome replication / viral process / chromosome organization / viral penetration into host nucleus / viral capsid ...viral genome packaging / T=16 icosahedral viral capsid / deNEDDylase activity / viral tegument / viral capsid assembly / viral DNA genome replication / viral process / chromosome organization / viral penetration into host nucleus / viral capsid / host cell / symbiont-mediated perturbation of host ubiquitin-like protein modification / cysteine-type deubiquitinase activity / host cell cytoplasm / symbiont entry into host cell / host cell nucleus / structural molecule activity / proteolysis / DNA binding Similarity search - Function | |||||||||||||||||||||
| Biological species |   Human herpesvirus 1 strain KOS Human herpesvirus 1 strain KOS | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||||||||||||||
 Authors Authors | Dai XH / Zhou ZH | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2018 Journal: Science / Year: 2018Title: Structure of the herpes simplex virus 1 capsid with associated tegument protein complexes. Authors: Xinghong Dai / Z Hong Zhou /  Abstract: Herpes simplex viruses (HSVs) rely on capsid-associated tegument complex (CATC) for long-range axonal transport of their genome-containing capsids between sites of infection and neuronal cell bodies. ...Herpes simplex viruses (HSVs) rely on capsid-associated tegument complex (CATC) for long-range axonal transport of their genome-containing capsids between sites of infection and neuronal cell bodies. Here we report cryo-electron microscopy structures of the HSV-1 capsid with CATC up to 3.5-angstrom resolution and atomic models of multiple conformers of capsid proteins VP5, VP19c, VP23, and VP26 and tegument proteins pUL17, pUL25, and pUL36. Crowning every capsid vertex are five copies of heteropentameric CATC, each containing a pUL17 monomer supporting the coiled-coil helix bundle of a pUL25 dimer and a pUL36 dimer, thus positioning their flexible domains for potential involvement in nuclear capsid egress and axonal capsid transport. Notwithstanding newly discovered fold conservation between triplex proteins and bacteriophage λ protein gpD and the previously recognized bacteriophage HK97 gp5-like fold in VP5, HSV-1 capsid proteins exhibit extraordinary diversity in forms of domain insertion and conformational polymorphism, not only for interactions with tegument proteins but also for encapsulation of large genomes. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7473.map.gz emd_7473.map.gz | 201.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7473-v30.xml emd-7473-v30.xml emd-7473.xml emd-7473.xml | 28.7 KB 28.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7473.png emd_7473.png | 290.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7473 http://ftp.pdbj.org/pub/emdb/structures/EMD-7473 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7473 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7473 | HTTPS FTP |
-Validation report
| Summary document |  emd_7473_validation.pdf.gz emd_7473_validation.pdf.gz | 77.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7473_full_validation.pdf.gz emd_7473_full_validation.pdf.gz | 77 KB | Display | |
| Data in XML |  emd_7473_validation.xml.gz emd_7473_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7473 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7473 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7473 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7473 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7473.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7473.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subparticle refinement of HSV-1 capsid vertex region with associated tegument proteins. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
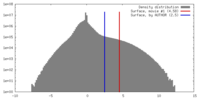
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human herpesvirus 1 strain KOS
| Entire | Name:   Human herpesvirus 1 strain KOS Human herpesvirus 1 strain KOS |
|---|---|
| Components |
|
-Supramolecule #1: Human herpesvirus 1 strain KOS
| Supramolecule | Name: Human herpesvirus 1 strain KOS / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Cultured in Vero cells. / NCBI-ID: 10306 / Sci species name: Human herpesvirus 1 strain KOS / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 200 MDa |
| Virus shell | Shell ID: 1 / Name: capsid / Diameter: 1300.0 Å / T number (triangulation number): 16 |
-Macromolecule #1: major capsid protein (MCP, VP5)
| Macromolecule | Name: major capsid protein (MCP, VP5) / type: protein_or_peptide / ID: 1 / Details: MCP forms the hexons and pentons of the capsid. / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 1 strain KOS / Strain: KOS Human herpesvirus 1 strain KOS / Strain: KOS |
| Sequence | String: MAAPNRDPPG YRYAAAMVPT GSLLSTIEVA SHRRLFDFFS RVRS DANSL YDVEFDALLG SYCNTLSLVR FLELGLSVAC VCTKFPELAY MNEGRVQFEV HQP LIARDG PHPIEQPTHN YMTKIIDRRA LNAAFSLATE AIALLTGEAL DGTGIGAHRQ LR AIQQLAR ...String: MAAPNRDPPG YRYAAAMVPT GSLLSTIEVA SHRRLFDFFS RVRS DANSL YDVEFDALLG SYCNTLSLVR FLELGLSVAC VCTKFPELAY MNEGRVQFEV HQP LIARDG PHPIEQPTHN YMTKIIDRRA LNAAFSLATE AIALLTGEAL DGTGIGAHRQ LR AIQQLAR NVQAVLGAFE RGTADQMLHV LLEKAPPLAL LLPMQRYLDN GRLATRVARA T LVAELKRS FCETSFFLGK AGHRREAVEA WLVDLTTATQ PSVAVPRLTH ADTRGRPVDG VLVTTAPIK QRLLQSFLKV EDTEADVPVT YGEMVLNGAN LVTALVMGKA VRSLDDVGR HLLEMQEEQL DLNRQTLDEL ESAPQTTRVR ADLVSIGEKL VFLEALEKRI YAATNVPY P LVGAMDLTFV LPLGLFNPVM ERFAAHAGDL VPAPGHPDPR AFPPRQLFFW GKDRQVL RL SLEHAIGTVC HPSLMNVDAA VGGLNRDPVE AANPYGAYVA APAGPAADMQ QLFLNA WGQ RLAHGRVRWV AEGQMTPEQF MQPDNANLAL ELHPAFDFFV GVADVELPGG DVPPA GPGE IQATWRVVNG NLPLALCPAA FRDARGLELG VGRHAMAPAT IAAVRGAFDD RNYP AVFYL LQAAIHGSEH VFCALARLVV QCITSYWNNT RCAAFVNDYS LVSYVVTYLG GDL PEECMA VYRDLVAHVE ALAQLVDDFT LTGPELGGQA QAELNHLMRD PALLPPLVWD CD ALMRRAA LDRHRDCRVS AGGHDPVYAA ACNVATADFN RNDGQLLHNT QARAADAADD R PHRGADWT VHHKIYYYVM VPAFSRGRCC TAGVRFDRVY ATLQNMVVPE IAPGEECPSD PVTDPAHPL HPANLVANTV NAMFHNGRVV VDGPAMLTLQ VLAHNMAERT TALLCSAAP DAGANTASTT NMRIFDGALH AGILLMAPQH LDHTIQNGDY FYPLPVHALF AGADHVAN A PNFPPALRDL SRQVPLVPPA LGANYFSSIR QPVVQHVRES AAGENALTYA LMAGYFK IS PVALHHQLKT GLHPGFGFTV VRQDRFVTEN VLFSERASEA YFLGQLQVAR HETGGG VNF TLTQPRGNVD LGVGYTAVVA TATVRNPVTD MGNLPQNFYL GRGAPPLLDN AAAVY LRNA VVAGNRLGPA QPVPVFGCAQ VPRRAGMDHG QDAVCEFIAT PVSTDVNYFR RPCN PRGRA AGGVYAGDKE GDVTALMYDH GQSDPSRAFA ATANPWASQR FSYGDLLYNG AYH LNGASP VLSPCFKFFT SADIAAKHRC LERLIVETGS AVSTATAASD VQFKRPPGCR EL VEDPCGL FQEAYPLTCA SDPALLRSAR NGEAHARETH FAQYLVYDAS PLKGLAL |
-Macromolecule #2: small capsid protein (SCP, VP26)
| Macromolecule | Name: small capsid protein (SCP, VP26) / type: protein_or_peptide / ID: 2 / Details: SCP binds on top of the MCP in hexons. / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 1 strain KOS / Strain: KOS Human herpesvirus 1 strain KOS / Strain: KOS |
| Sequence | String: MAVPQFHRPS TVTTDSVRAL GMRGLVLATN NSQFIMDNNH PHPQ GTQGA VREFLRGQAA ALTDLGLAHA NNTFTPQPMF AGDAPAAWLR PAFGLRRTYS PFV VREPST PGTP |
-Macromolecule #3: capsid triplex subunit 1 (VP19C)
| Macromolecule | Name: capsid triplex subunit 1 (VP19C) / type: protein_or_peptide / ID: 3 Details: Complexed 1:2 with capsid triplex subunit 2 to form triplexes on the capsid shell. Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 1 strain KOS / Strain: KOS Human herpesvirus 1 strain KOS / Strain: KOS |
| Sequence | String: MKTNPLPATP SVWGGSTVEL PPTTRDTAGQ GLLRRVLRPP ISRR DGPGL PRGSGPRRAA STLWLLGLDG TDAPPGALTP NDDTEQALDK ILRGTMRGGA ALI GSPRHH LTRQVILTDL CQPNADRAGT LLLALRHPAD LPHLAHQRAP PGRQTERLGE AW GQLMEAT ...String: MKTNPLPATP SVWGGSTVEL PPTTRDTAGQ GLLRRVLRPP ISRR DGPGL PRGSGPRRAA STLWLLGLDG TDAPPGALTP NDDTEQALDK ILRGTMRGGA ALI GSPRHH LTRQVILTDL CQPNADRAGT LLLALRHPAD LPHLAHQRAP PGRQTERLGE AW GQLMEAT ALGSGRAESG CTRAGLVSFN FLVAACAASY DARDAADAVR AHVTANYRGT R VGARLDRF SECLRAMVHT HVFPHEVMRF FGGLVSWVTQ DELASVTAVC AGPQEAAHTG HPGRPRSAV ILPACAFVDL DAELGLGGPG AAFLYLVFTY RQRRDQELCC VYVIKSQLP PRGLEPALER LFGRLRITNT IHGTEDMTPP APNRNPDFPL AGLAANPQTP RCSAGQVT N PQFADRLYRW QPDLRGRPTA RTCTYAAFAE LGMMPEDSPR CLHRTERFGA VSVPVVI LE GVVWRPGEWR ACA |
-Macromolecule #4: capsid triplex subunit 2 (VP23)
| Macromolecule | Name: capsid triplex subunit 2 (VP23) / type: protein_or_peptide / ID: 4 Details: Complexed 2:1 with capsid triplex subunit 1 to form triplexes on the capsid shell. Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 1 strain KOS / Strain: KOS Human herpesvirus 1 strain KOS / Strain: KOS |
| Sequence | String: MLADGFETDI AIPSGISRPD AAALQRCEGR VVFLPTIRRQ LTLA DVAHE SFVSGGVSPD TLGLLLAYRR RFPAVITRVL PTRIVACPLD VGLTHAGTVN LRN TSPVDL CNGDPISLVP PVFEGQATDV RLDSLDLTLR FPVPLPSPLA REIVARLVAR GI RDLNPSP ...String: MLADGFETDI AIPSGISRPD AAALQRCEGR VVFLPTIRRQ LTLA DVAHE SFVSGGVSPD TLGLLLAYRR RFPAVITRVL PTRIVACPLD VGLTHAGTVN LRN TSPVDL CNGDPISLVP PVFEGQATDV RLDSLDLTLR FPVPLPSPLA REIVARLVAR GI RDLNPSP RNPGGLPDLN VLYYNGSRLS LLADVQQLGP VNAELRSLVL NMVYSITEGT T IILTLIPR LFALSAQDGY VNALLQMQSV TREAAQLIHP EAPALMQDGE RRLPLYEALV AWLTHAGQL GDTLALAPVV RVCTFDGAAV VRSGDMAPVI RYP |
-Macromolecule #5: tegument protein pUL17
| Macromolecule | Name: tegument protein pUL17 / type: protein_or_peptide / ID: 5 Details: Component of the capsid-associated tegument complex (CATC). Each CATC contains one pUL17 subunit. Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 1 strain KOS / Strain: KOS Human herpesvirus 1 strain KOS / Strain: KOS |
| Sequence | String: MNAHLANEVQ YDLGHGPGRP SSLVHVIISS ECLAAAGIPL AALM RGRPG LGTAANFQVE IQTRAHATGD CTPWCTAFAA YVPADAVGEL LAPVVPAHPG LLP RASSAG GLFVSLPVVC DAQGVYDPYA VAALRLAWGS GASCARVILF SYDELVPPNT RY AADSTRI ...String: MNAHLANEVQ YDLGHGPGRP SSLVHVIISS ECLAAAGIPL AALM RGRPG LGTAANFQVE IQTRAHATGD CTPWCTAFAA YVPADAVGEL LAPVVPAHPG LLP RASSAG GLFVSLPVVC DAQGVYDPYA VAALRLAWGS GASCARVILF SYDELVPPNT RY AADSTRI MRVCRHLCRY VALLGAAAPP AAKEAAAHLS MGLGESASPR PQPLARPHAG A PADPPIVG ASDPPISPEE QLTAPGGDTT AAQDVSIAQE NEEILALVQR AVQDVTRRHP VRARTGRAA CGVASGLRQG ALVHQAVSGG AMGAADADAV LAGLEPPGGG RFVAPAPHG PGGEDILNDV LTLTPGTAKP RSLVEWLDRG WEALAGGDRP DWLWSRRSIS VVLRHHYG T KQRFVVVSYE NSVAWGGRRA RPPLLSSALA TALTEACAAE RVVRPHQLSP AGQAELL LR FPALEVPLRH PRPVLPPFDI AAEVAFTARI HLACLRALGQ AIRAALQGGP RISQRL RYD FGPDQRAWLG EVTRRFPILL ENLMRAVEGT APDAFFHTAY ALAVLAHLGG RGGRG RRVV PLGDDLPARF ADSDGHYVFD YYSTSGDTLR LNNRPIAVAM DGDVSKREQS KCRF MEAVP STAPRRVCEQ YLPGESYAYL CLGFNRRLCG IVVFPGGFAF TINIAAYLSL SDP VARAAV LRFCRKVSSG NGRSR |
-Macromolecule #6: tegument protein pUL25
| Macromolecule | Name: tegument protein pUL25 / type: protein_or_peptide / ID: 6 Details: Component of the capsid-associated tegument complex (CATC). Each CATC contains two pUL25 subunits. Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 1 strain KOS / Strain: KOS Human herpesvirus 1 strain KOS / Strain: KOS |
| Sequence | String: MDPYCPFDAL DVWEHRRFIV ADSRNFITPE FPRDFWMSPV FNLP RETAA EQVVVLQAQR TAAAAALENA AMQAAELPVD IERRLRPIER NVHEIAGALE ALE TAAAAA EEADAARGDE PAGGGDGGAP PGLAVAEMEV QIVRNDPPLR YDTNLPVDLL HM VYAGRGA ...String: MDPYCPFDAL DVWEHRRFIV ADSRNFITPE FPRDFWMSPV FNLP RETAA EQVVVLQAQR TAAAAALENA AMQAAELPVD IERRLRPIER NVHEIAGALE ALE TAAAAA EEADAARGDE PAGGGDGGAP PGLAVAEMEV QIVRNDPPLR YDTNLPVDLL HM VYAGRGA TGSSGVVFGT WYRTIQDRTI TDFPLTTRSA DFRDGRMSKT FMTALVLSLQ A CGRLYVGQ RHYSAFECAV LCLYLLYRNT HGAADDSDRA PVTFGDLLGR LPRYLACLAA VIGTEGGRP QYRYRDDKLP KTQFAAGGGR YEHGALASHI VIATLMHHGV LPAAPGDVP RDASTHVNPD GVAHHDDINR AAAAFLSRGH NLFLWEDQTL LRATANTITA LGVIQRLL A NGNVYADRLN NRLQLGMLIP GAVPSEAIAR GASGSDSGAI KSGDNNLEAL CANYVLP LY RADPAVELTQ LFPGLAALCL DAQAGRPVGS TRRVVDMSSG ARQAALVRLT ALELIN RTR TNPTPVGEVI HAHDALAIQY EQGLGLLAQQ ARIGLGSNTK RFSAFNVSSD YDMLY FLCL GFIPQYLSAV |
-Macromolecule #7: tegument protein pUL36
| Macromolecule | Name: tegument protein pUL36 / type: protein_or_peptide / ID: 7 Details: Component of the capsid-associated tegument complex (CATC). Each CATC contains two pUL36 subunits. Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 1 strain KOS / Strain: KOS Human herpesvirus 1 strain KOS / Strain: KOS |
| Sequence | String: MIAGTPPHST MERGGDRDIV VTGARNQFAP DLEPGGSVSC MRSS LSFLS LIFDVGPRDV LSAEAIEGCL VEGGEWTRAT AGPGPPRMCS IVELPNFLEY PGA RGGLRC VFSRVYGEVG FFGEPAAGLL ETQCPAHTFF AGPWALRPLS YTLLTIGPLG MG LFRDGDT ...String: MIAGTPPHST MERGGDRDIV VTGARNQFAP DLEPGGSVSC MRSS LSFLS LIFDVGPRDV LSAEAIEGCL VEGGEWTRAT AGPGPPRMCS IVELPNFLEY PGA RGGLRC VFSRVYGEVG FFGEPAAGLL ETQCPAHTFF AGPWALRPLS YTLLTIGPLG MG LFRDGDT AYLFDPHGLP EGTPAFIAKV RAGDMYPYLT YYTRDRPDVR WAGAMVFFVP S GPEPAAPA DLTAAALHLY GASETYLQDE AFSERRVAIT HPLRGEIAGL GEPCVGVGPR EGVGGPGPH PPTAAQSPPP TRARRDDRAS ETSRGTAGPS AKPEAKRPNR APDDVWAVA LKGTPPTDPP SADPPSADPP SAIPPPPPSA PKTPAAEAAE EDDDDMRVLE MGVVPVGR H RARYSAGLPK RRRPTWTPPS SVEDLTSGEK TKRSAPPAKT KKKSTPKGKT PVGAAVP AS VPEPVLASAP PDPAGPPVAE AGEDDGPTVP ASSQALEALK TRRSPEPPGA DLAQLF EAH PNVAATAVKF TACSAALARE VAACSRLTIS ALRSPYPASP GLLELCVIFF FERVL AFLI ENGARTHTQA GVAGPAAALL EFTLNMLPWK TAVGDFLAST RLSLADVAAH LPLV QHVLD ENSLIGRLAL AKLILVARDV IRETDAFYGE LADLELQLRA APPANLYTRL GEW LLERSQ AHPDTLFAPA TPTHPEPLLY RVQALAKFAR GEEIRVEAED RQMREALDAL AR GVDAVSQ HAGPLGVMPA PAGAAPQGAP RPPPLGPEAV QVRLEEVRTQ ARRAIEGAVK E YFYRGAVY SAKALQASDN NDRRFHVASA AVVPVVQLLE SLPVFDQHTR DIAQRAAIPA PPPIATSPT AILLRDLIQR GQTLDAPEDL AAWLSVLTDA ANQGLIERKP LDELARSIR DINDQQARRS SGLAELRRFD ALDAALGQQL DSDAAFVPAP GASPYPDDGG LSPEATRM A EEALRQARAM DAAKLTAELA PDARARLRER ARSLEAMLEG ARERAKVARD AREKFLH KL QGVLRPLPDF VGLKACPAVL ATLRASLPAG WSDLPEAVRG APPEVTAALR ADMWGL LGQ YRDALEHPTP DTATALSGLH PSFVVVLKNL FADAPETPFL LQFFADHAPI IAHAV SNAI NAGSAAVATA DPASTVDAAV RAHRVLVDAV TALGAAASDP ASPLAFLAAM ADSA AGYVK ATRLALDARV AIAQLTTLGS AAADLVVQVR RAANQPEGEH ASLIQAATRA TTG ARESLA GHEGRFGGLL HAEGTAGDHS PSGRALQELG KVIGATRRRA DELEAATADL RE KMAAQRA RSSHERWAAD VEAVLDRVES GAEFDVVELR RLQALAGTHG YNPRDFRKRA E QALGTNAK AVTLALETAL AFNPYTPENQ RHPMLPPLAA IHRIDWSAAF GAAADTYADM FRVDTEPLA RLLRLAGGLL ERAQANDGFI DYHEAVLHLS EDLGGVPALR QYVPFFQKG YAEYVDIRDR LDALRADARR AIGSVALDLA AAAEEISAVR NDPAAAAELV RAGVTLPC P SEDALVACVA ALERVDQSPV KDTAYADYVA FVTRQDLADT KDAVVRAKQQ RAEATER VT AGLREVLAAR ERRAQLEAEG LANLKTLLKV VAVPATVAKT LDQARSAEEI ADQVEI LVD QTEKARELDV QAVAWLEHAQ RTFETHPLSA ASGDGPGLLT RQGARLQALF DTRRR VEAL RRSLEEAEAE WDEVWGRFGR VRGGAWKSPE GFRAACEQLR ALQDTTNTVS GLRA QRDYE RLPAKYQGVL GAKSAERAGA VEELGGRVAQ HADLSARLRD EVVPRVAWEM NFD TLGGLL AEFDAVAGDL APWAVEEFRG ARELIQRRMG LYSAYAKATG QTGAGAAAAP AP LLVDLRA LDARARASAP PGQEADPQML RRRGEAYLRV SGGPGPLVLR EATSTLDRPF A PSFLVPDG TPLQYALCFP AVTDKLGALL MCPEAACIRP PLPTDTLESA STVTAMYVLT VINRLQLAL SDAQAANFQL FGRFVRHRQA RWGASMDAAA ELYVALVATT LTREFGCRW AQLEWGGDAA APGPPLGPQS STRHRVSFNE NDVLVALVAS SPEHIYTFWR LDLVRQHE Y MHLTLPRAFQ NAADSMLFVQ RLTPHPDARI RVLPAFSAGG PPTRGLMFGT RLADWRR GK LSETDPLAPW RSVPELGTER GAALGKLSPA QALAAVSVLG RMCLPSTALV ALWTCM FPD DYTEYDSFDA LLTARLESGQ TLSPSGGREA SPPAPPNALY RPTGQHVAVP AAATH RTPA ARVTAMDLVL AAVLLGAPVV VALRNTTAFS RESELELCLT LFDSRARGPD AALR DAVSS DIETWAVRLL HADLNPIENA CLAAQLPRLS ALIAERPLAR GPPCLVLVDI SMT PVAVLW ENPDPPGPPD VRFVGSEATE ELPFVAGGED VLAASATDED PFLARAILGR PF DASLLSG ELFPGHPVYQ RAPDDQSPSV PNPTPGPVDL VGAEGSLGPG SLAPTLFTDA T PGEPVPPR MWAWIHGLEE LASDDSGGPA PLLAPDPLSP TADQSVPTSQ CAPRPPGPAV TAREARPGV PAESTRPAPV GPRDDFRRLP SPQSSPAPPD ATAPRPPASS RASAASSSG SRARRHRRAR SLARATQASA TTQGWRPPAL PDTVAPVTDF ARPPAPPKPP EPAPHALV S GVPLPLGPQA AGQASPALPI DPVPPPVATG TVLPGGENRR PPLTSGPAPT PPRVPVG GP QRRLTRPAVA SLSESRESLP SPWDPADPTA PVLGRNPAEP TSSSPAGPSP PPPAVQ PVA PPPTSGPPPT YLTLEGGVAP GGPVSRRPTT RQPVATPTTS ARPRGHLTVS RLSAP QPQP QPQPQPQPQP QPQPQPQPQP QPQPQPQPQP QPQPQPQPQP QPQPQPQPQP QPQP QPQPQ PQPQPQNGHV APGEYPAVRF RAPQNRPSVP ASASSTNPRT GSSLSGVSSW ASS LALHID ATPPPVSLLQ TLYVSDDEDS DATSLFLSDS EAEALDPLPG EPHSPITNEP FS ALSADDS QEVTRLQFGP PPVSANAVLS RRYVQRTGRS ALAVLIRACY RLQQQLQRTR R ALLHHSDA VLTSLHHVRM LLG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Component - Formula: PBS / Component - Name: phosphate buffered saline |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 298 K / Instrument: HOMEMADE PLUNGER Details: The sample was manually blotted and frozen with a homemade plunger.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 79.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 7676 pixel / Digitization - Dimensions - Height: 7420 pixel / Digitization - Sampling interval: 2.5 µm / Digitization - Frames/image: 1-26 / Number grids imaged: 3 / Number real images: 7356 / Average exposure time: 13.0 sec. / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 24271 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 14000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)