+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7450 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Gasdermin A3 membrane pore | |||||||||
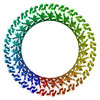
 Map data Map data | Gasdermin A3 pore with 28-fold symmetry | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
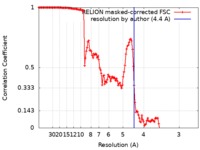
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Ruan J / Xia S / Wu H | |||||||||
 Citation Citation |  Journal: Nature / Year: 2018 Journal: Nature / Year: 2018Title: Cryo-EM structure of the gasdermin A3 membrane pore. Authors: Jianbin Ruan / Shiyu Xia / Xing Liu / Judy Lieberman / Hao Wu /  Abstract: Gasdermins mediate inflammatory cell death after cleavage by caspases or other, unknown enzymes. The cleaved N-terminal fragments bind to acidic membrane lipids to form pores, but the mechanism of ...Gasdermins mediate inflammatory cell death after cleavage by caspases or other, unknown enzymes. The cleaved N-terminal fragments bind to acidic membrane lipids to form pores, but the mechanism of pore formation remains unresolved. Here we present the cryo-electron microscopy structures of the 27-fold and 28-fold single-ring pores formed by the N-terminal fragment of mouse GSDMA3 (GSDMA3-NT) at 3.8 and 4.2 Å resolutions, and of a double-ring pore at 4.6 Å resolution. In the 27-fold pore, a 108-stranded anti-parallel β-barrel is formed by two β-hairpins from each subunit capped by a globular domain. We identify a positively charged helix that interacts with the acidic lipid cardiolipin. GSDMA3-NT undergoes radical conformational changes upon membrane insertion to form long, membrane-spanning β-strands. We also observe an unexpected additional symmetric ring of GSDMA3-NT subunits that does not insert into the membrane in the double-ring pore, which may represent a pre-pore state of GSDMA3-NT. These structures provide a basis that explains the activities of several mutant gasdermins, including defective mutants that are associated with cancer. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7450.map.gz emd_7450.map.gz | 9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7450-v30.xml emd-7450-v30.xml emd-7450.xml emd-7450.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7450_fsc.xml emd_7450_fsc.xml | 13.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_7450.png emd_7450.png | 82.5 KB | ||
| Others |  emd_7450_half_map_1.map.gz emd_7450_half_map_1.map.gz emd_7450_half_map_2.map.gz emd_7450_half_map_2.map.gz | 108.6 MB 108.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7450 http://ftp.pdbj.org/pub/emdb/structures/EMD-7450 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7450 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7450 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7450.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7450.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Gasdermin A3 pore with 28-fold symmetry | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half map 1 of the Gasdermin A3 pore with 28-fold symmetry
| File | emd_7450_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 of the Gasdermin A3 pore with 28-fold symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 of the Gasdermin A3 pore with 28-fold symmetry
| File | emd_7450_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 of the Gasdermin A3 pore with 28-fold symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Gasdermin A3 N-terminal domain
| Entire | Name: Gasdermin A3 N-terminal domain |
|---|---|
| Components |
|
-Supramolecule #1: Gasdermin A3 N-terminal domain
| Supramolecule | Name: Gasdermin A3 N-terminal domain / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: mouse Gasdermin A3 pores with 28-fold symmetry |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 810 KDa |
-Macromolecule #1: Gasdermin A3
| Macromolecule | Name: Gasdermin A3 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MHMPVFEDVT RALVRELNPR GDLTPLDSLI DFKHFRPFCL VLRKRKSTLF WGARYVRTDY TLLDLLEPGS SPSDLTDSGN FSFKNMLDVQ VQGLVEVPKT VKVKGTAGLS QSSTLEVQTL SVAPSALENL KKERKLSADH SFLNEMRYHE KNLYVVMEAV EAKQEVTVEQ ...String: MHMPVFEDVT RALVRELNPR GDLTPLDSLI DFKHFRPFCL VLRKRKSTLF WGARYVRTDY TLLDLLEPGS SPSDLTDSGN FSFKNMLDVQ VQGLVEVPKT VKVKGTAGLS QSSTLEVQTL SVAPSALENL KKERKLSADH SFLNEMRYHE KNLYVVMEAV EAKQEVTVEQ TGNANAIFSL PSLALLGLQG SLNNNKAVTI PKGCVLAYRV RLLRVFLFNL WDIPYICNDS MQTFPKIRRV PCSAFISPTQ MISEEPEEEK LIGELEVLFQ |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)